Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. May 21, 2014; 20(19): 5708-5720
Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5708
Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5708
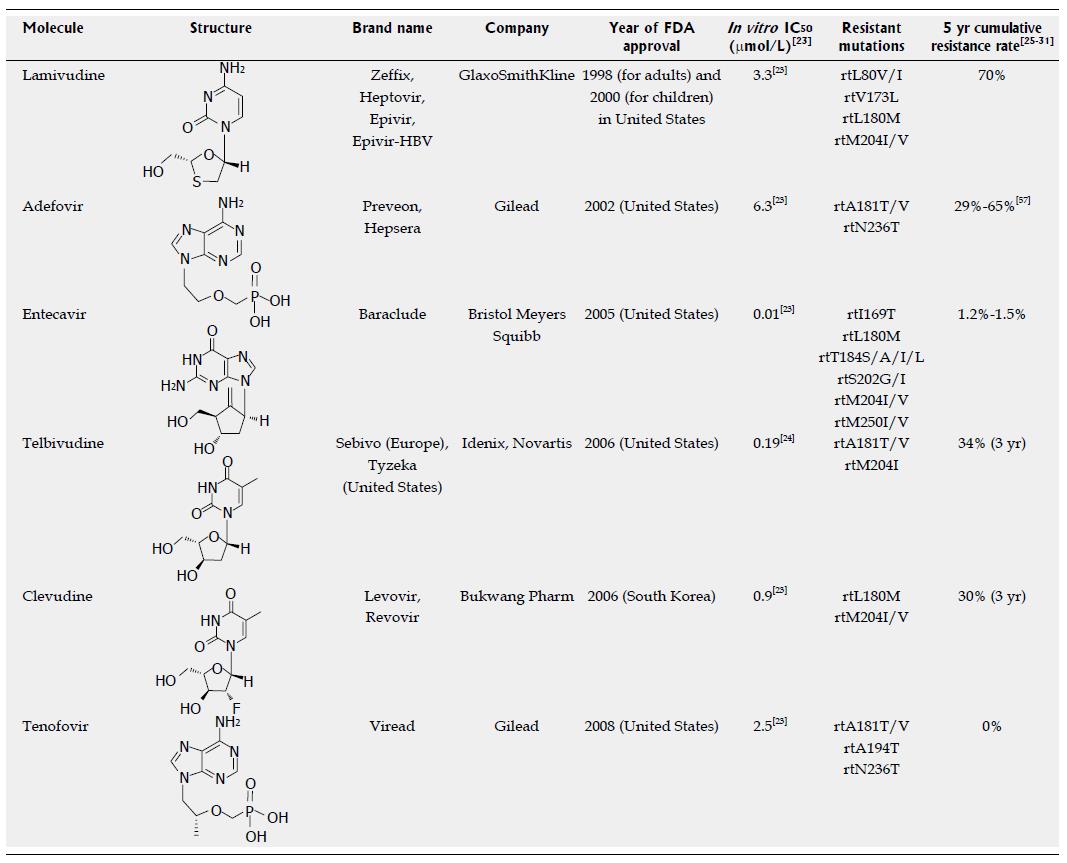
Figure 1 Approved anti-hepatitis B virus drugs and their resistant mutations.
IC50 values are dependent on the duration of drug exposure to cells, the cells used, and the protocol used.
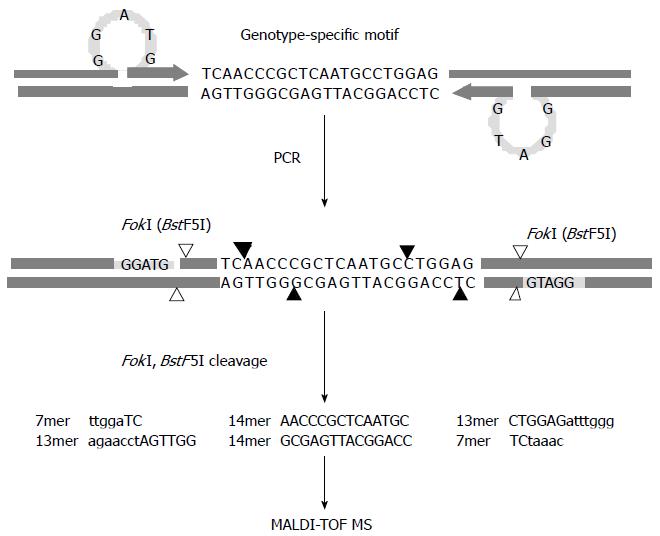
Figure 2 Schematic diagram of the restriction fragment mass polymorphism genotyping strategy.
Polymerase chain reaction is performed with primers designed to introduce a type IIS restriction endonuclease recognition sequence (FokI) ahead of the genotype-specific motifs upon amplification. The enzymatic cleavage of the products leads to excision of multiple oligonucleotide fragments representing the motifs shown in capital letters, and then the masses of the resulting oligonucleotide fragments are examined by MALDI-TOF MS. Cleavage sites of FokI and BstF5I, an isoschizomer of FokI, are indicated by filled and blank arrows, respectively, and recognition sites for both restriction endonucleases are specified by the shaded bars.
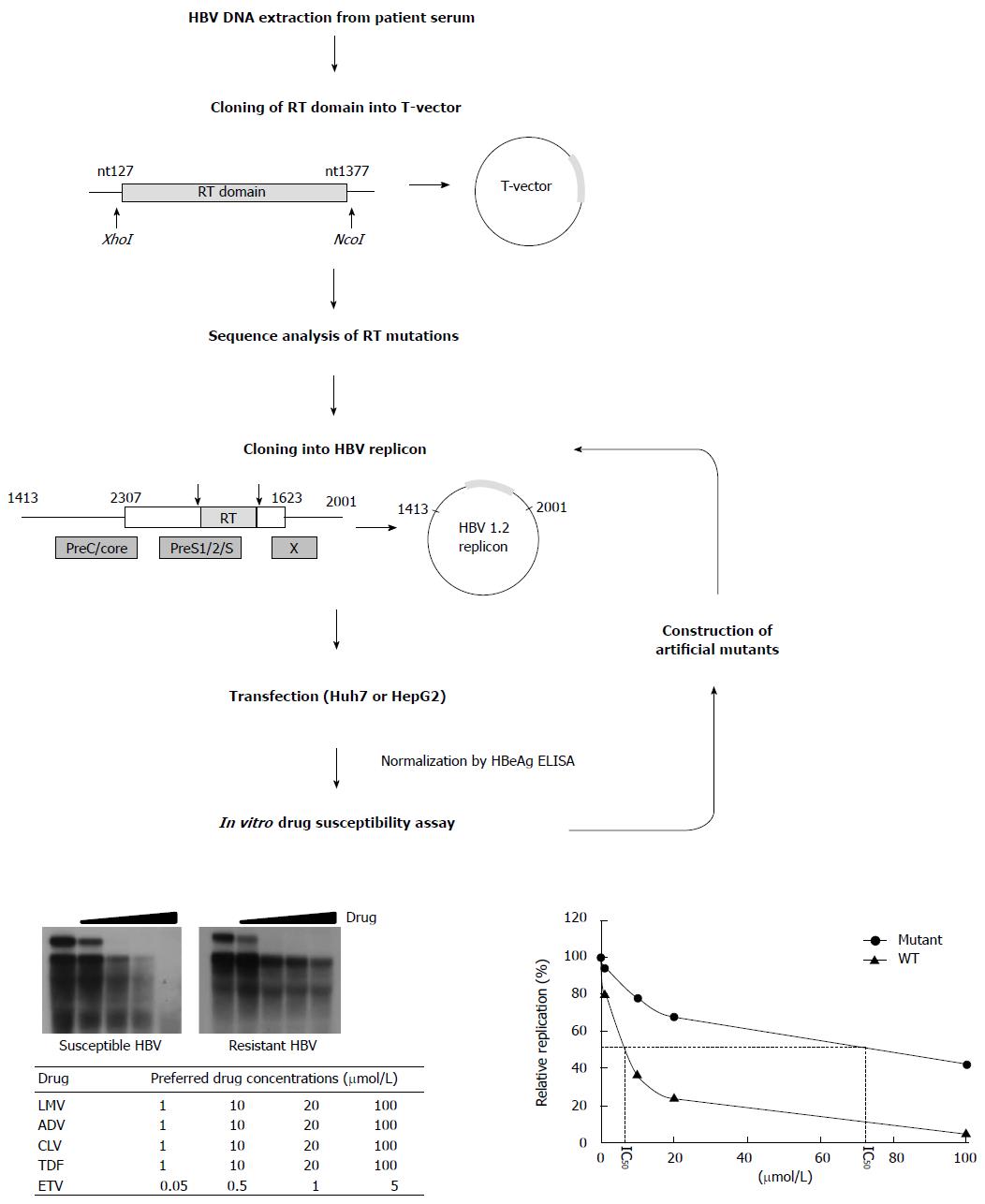
Figure 3 Scheme for in vitro phenotypic validation of drug-resistant hepatitis B virus.
Hepatitis B virus (HBV) DNA is purified from patient serum, and the sequence of RT mutations is analyzed. After cloning into replication-competent HBV replicons, each mutant is transfected into hepatoma cell lines followed by Southern blot (or real time polymerase chain reaction) analysis. The IC50 (μmol/L) value is obtained by quantification of replication ability and curve-fitting. To characterize the specific mutation(s) conferring resistance to antiviral drugs, each artificial mutant must be constructed and individually tested.
- Citation: Kim JH, Park YK, Park ES, Kim KH. Molecular diagnosis and treatment of drug-resistant hepatitis B virus. World J Gastroenterol 2014; 20(19): 5708-5720
- URL: https://www.wjgnet.com/1007-9327/full/v20/i19/5708.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i19.5708