Copyright
©2008 The WJG Press and Baishideng.
World J Gastroenterol. Jan 14, 2008; 14(2): 203-210
Published online Jan 14, 2008. doi: 10.3748/wjg.14.203
Published online Jan 14, 2008. doi: 10.3748/wjg.14.203
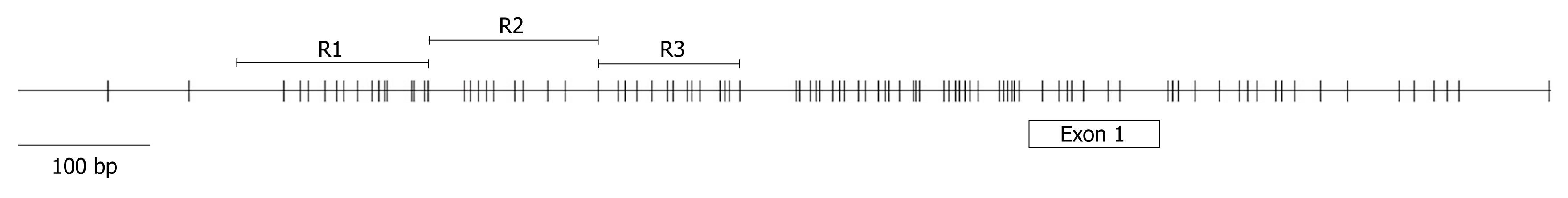
Figure 1 Schematic representation of the TIMP3 promoter.
The schematic was generated by downloading the TIMP3 CpG island genetic element from http://genome.ucsc.edu/, using the Human March 2006 Freeze. The relative location of TIMP3 exon 1 is indicated. Each vertical line represents the location of a CpG. The three overlapping regions (R1, R2 and R3) amplified for methylation analysis are shown.
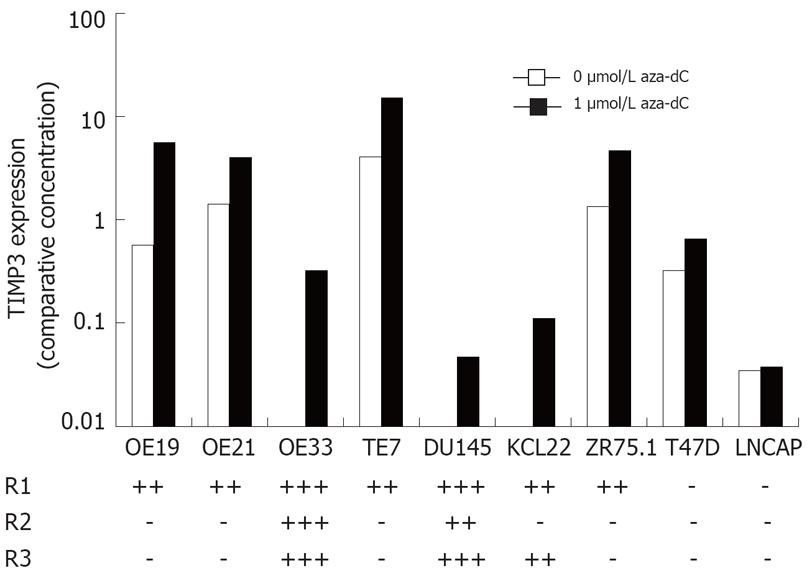
Figure 2 Expression and methylation of TIMP3 in cancer cell lines.
The level of TIMP3 mRNA in cell lines treated with either 0 or 1 &mgr;mol/L aza-dC was determined by qRT-PCR and normalized using PBGD. Methylation was determined by melt curve analysis in three overlapping regions R1, R2 and R3, and is summarized.
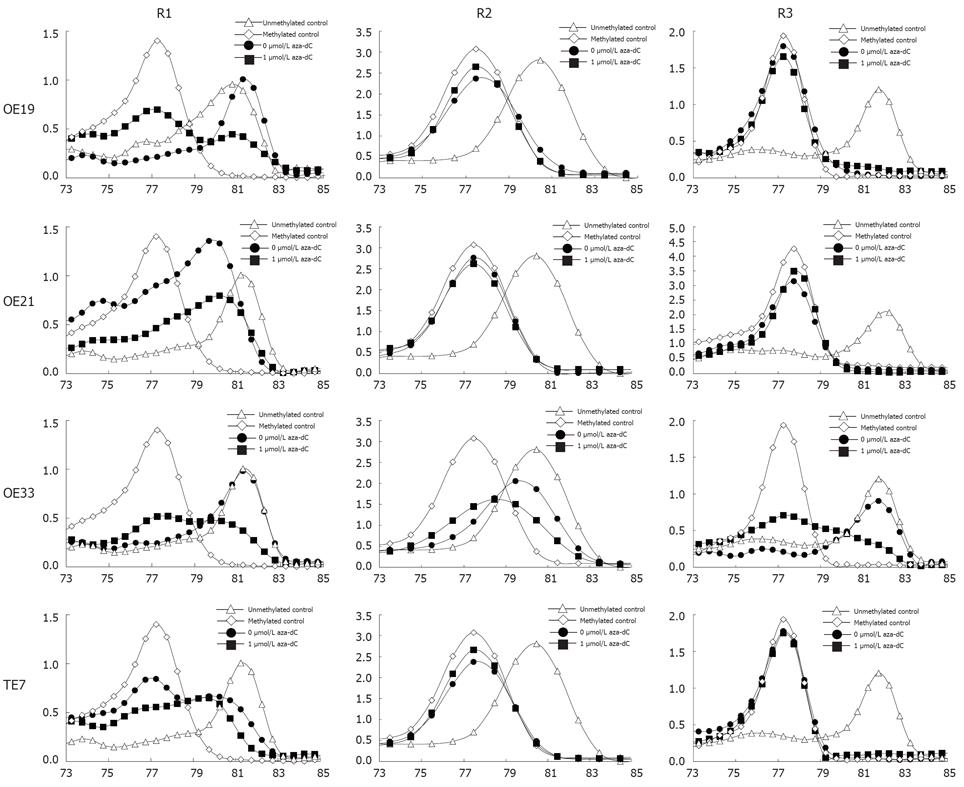
Figure 3 Methylation of TIMP3 in esophageal cancer cell lines.
The X axis is temperature, the Y axis is fluorescence dF/dT.
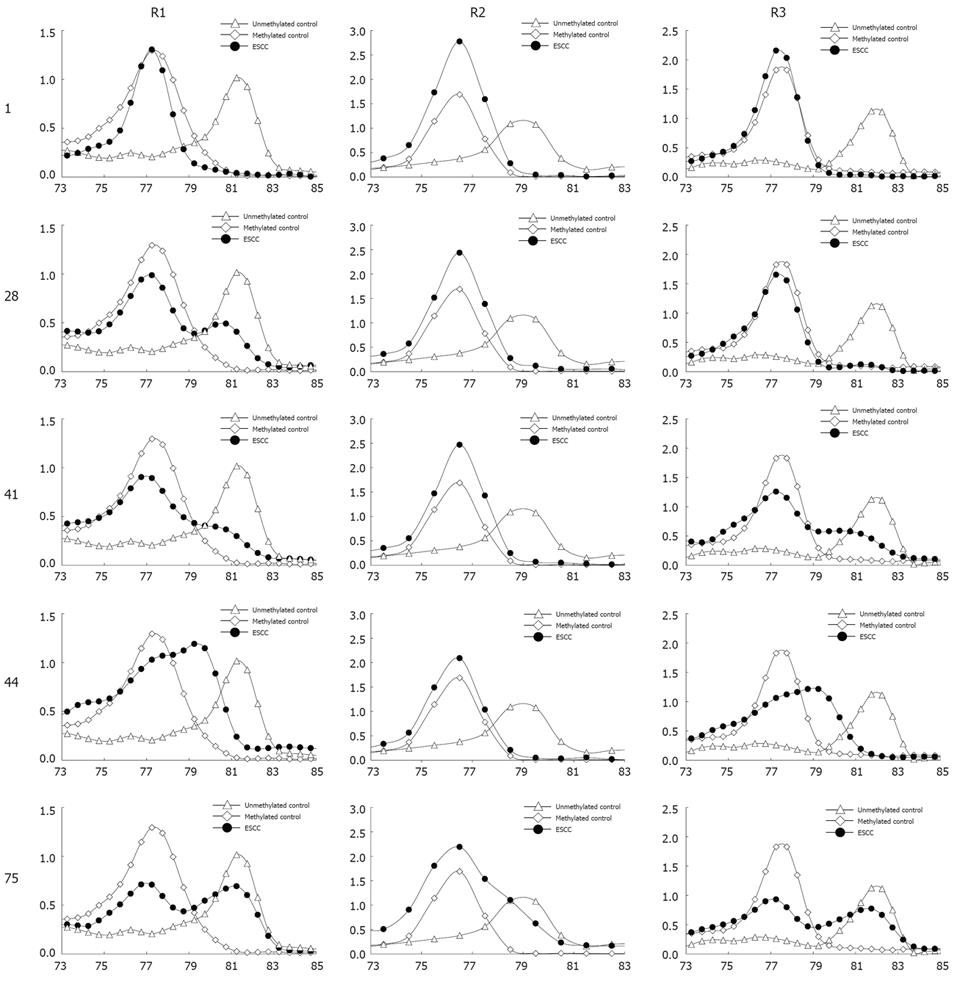
Figure 4 Methylation of TIMP3 in ESCC samples.
The X axis is temperature, the Y axis is fluorescence dF/dT.
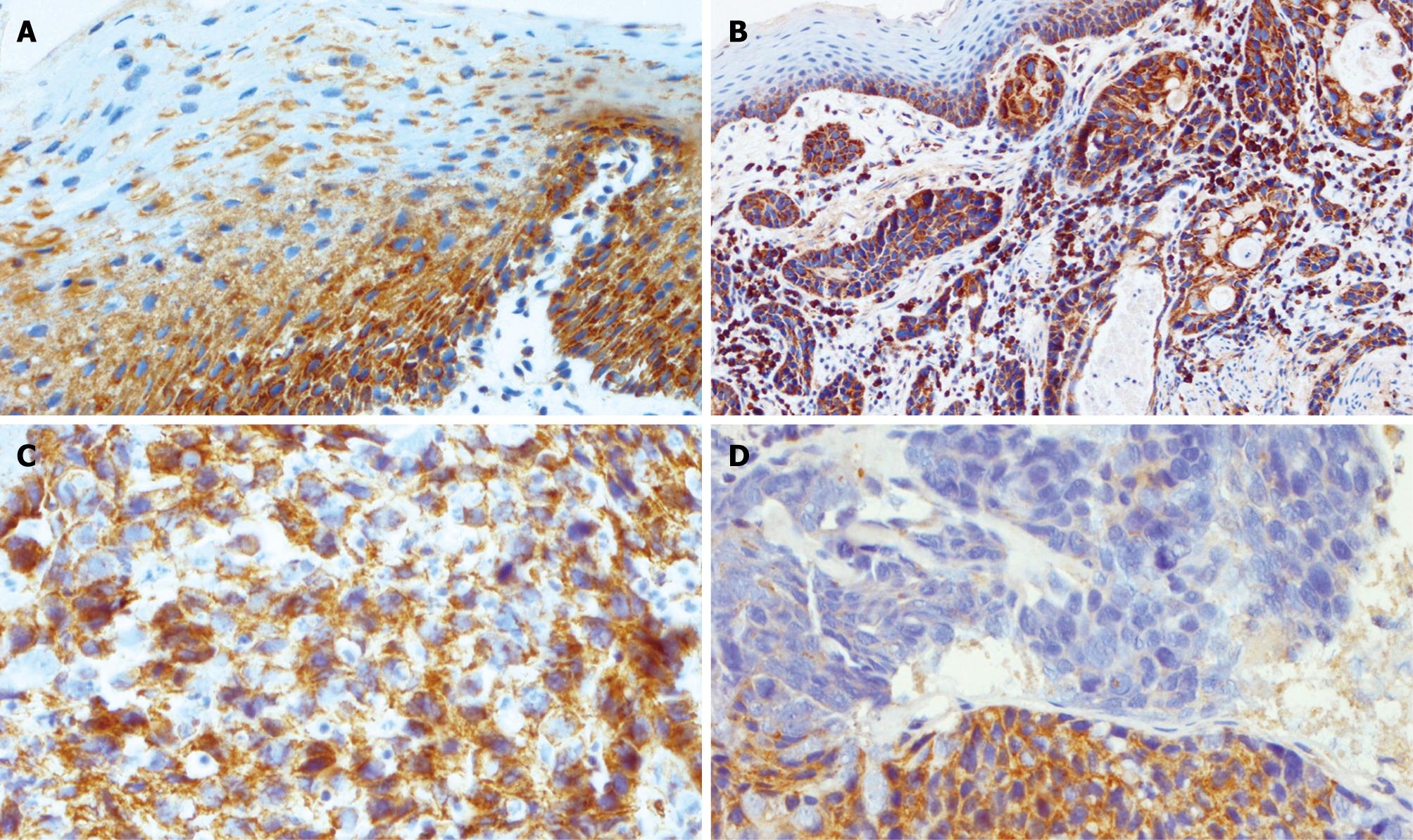
Figure 5 Expression of TIMP3 in esophageal tissues.
A: Strong heterogeneous cytoplasmic expression of TIMP3 was limited to the basal layers in normal squamous mucosa; B: TIMP3 expression in unmethylated ESCC underlying normal squamous mucosa; C: Unmethylated ESCC showing strong heterogeneous staining; D: Methylated ESCC showing the region of reduced expression.
- Citation: Smith E, Young NJD, Tian ZQ, Caruso M, Ruszkiewicz AR, Liu JF, Jamieson GG, Drew PA. Methylation of TIMP3 in esophageal squamous cell carcinoma. World J Gastroenterol 2008; 14(2): 203-210
- URL: https://www.wjgnet.com/1007-9327/full/v14/i2/203.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.203