©2007 Baishideng Publishing Group Inc.
World J Gastroenterol. Dec 14, 2007; 13(46): 6254-6258
Published online Dec 14, 2007. doi: 10.3748/wjg.v13.i46.6254
Published online Dec 14, 2007. doi: 10.3748/wjg.v13.i46.6254
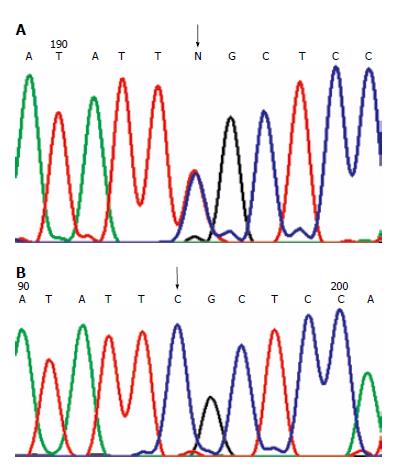
Figure 1 Missense mutations.
(A): MLH1 mutation in H2 family at 649 codon 217 exon 8: CGC→TGC. The arrow shows the site of the mutation; (B): Wild-type sequence. The arrow shows the corresponding site.
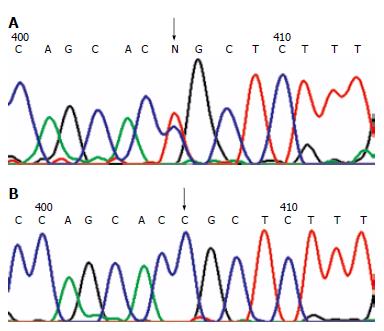
Figure 2 Missense mutation.
A: MLH1 mutation in H31 family at 1742 odon 581 exon 16: CCG→CTG. The arrow shows the site of the mutation; B: Wild-type sequence. The arrow shows the corresponding site.
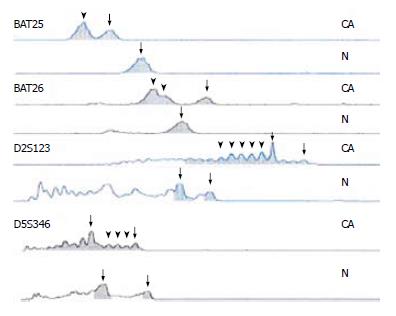
Figure 3 Four sites of MSI in H2 family.
CA represents the tumor tissue, N represents the control tissue, the arrow-heads show new waves, and the arrows show wild ones.
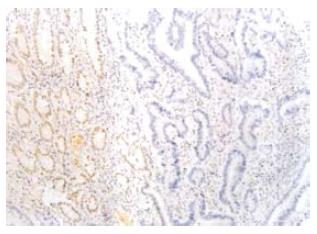
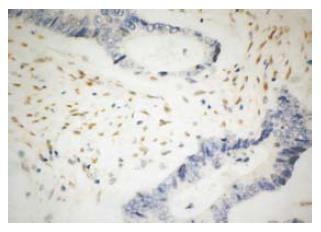
Figure 4 MLH1 protein in H2 was negative in the tumor glands (right), and positive in the mucous glands next to the tumor tissue (EnVison, × 200).
Figure 5 MLH1 protein in H31 was negative in the tumor glands, and positive in the stroma cells (EnVison, × 400).
- Citation: Wang CF, Zhou XY, Zhang TM, Xu Y, Cai SJ, Shi DR. Two novel germline mutations of MLH1 and investigation of their pathobiology in hereditary non-polyposis colorectal cancer families in China. World J Gastroenterol 2007; 13(46): 6254-6258
- URL: https://www.wjgnet.com/1007-9327/full/v13/i46/6254.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i46.6254