©2007 Baishideng Publishing Group Co.
World J Gastroenterol. Jan 7, 2007; 13(1): 48-64
Published online Jan 7, 2007. doi: 10.3748/wjg.v13.i1.48
Published online Jan 7, 2007. doi: 10.3748/wjg.v13.i1.48
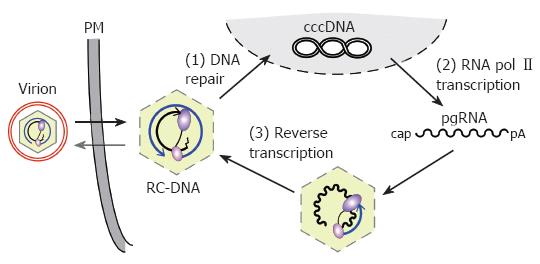
Figure 1 Replication cycle of the hepadnaviral genome.
Enveloped virions infect the cell, releasing RC-DNA containing nucleocapsids into the cytoplasm. RC-DNA is transported to the nucleus, and repaired to form cccDNA (1). Transcription of cccDNA by RNA polymerase II (2) produces, amongst other transcripts (not shown), pgRNA. pgRNA is encapsidated, together with P protein, and reverse transcribed inside the nucleocapsid (3). (+)-DNA synthesis from the (-)-DNA template generates new RC-DNA. New cycles lead to intracellular cccDNA amplification; alternatively, the RC-DNA containing nucleocapsids are enveloped and released as virions. PM, plasma membrane.
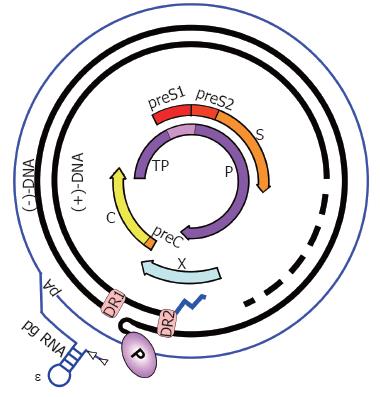
Figure 2 HBV genome organization.
The partially double-stranded, circular RC-DNA is indicated by thick black lines, with P covalently linked to the 5´ end of the (-)-DNA, and the RNA primer (zigzag line) at the 5´ end of (+)-DNA. The dashed part symbolizes the heterogeneous lengths of the (+)-strands. DR1 and DR2 are the direct repeats. The outer circle symbolizes the terminally redundant pgRNA with ε close to the 5´ end, and the poly-A tail at the 3´ end. The precore mRNA is nearly identical, except it starts slightly upstream. The relative positions of the open reading frames for core (C), P, preS/S, and X are shown inside. TP, Terminal protein domain of P.
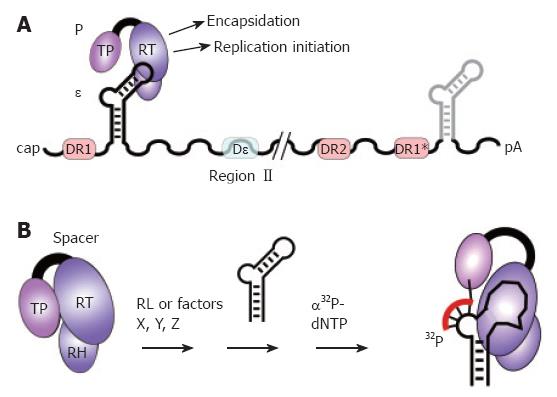
Figure 3 The pgRNA as substrate for P protein.
A: pgRNA organization. The pgRNA is shown with some major cis-elements, i.e. ε (hairpin structure), DR1, DR2, and DR1*; DHBV, but not HBV, requires for encapsidation an additional cis-element (Dε region II). Binding of P protein to 5´ ε, but not 3´ ε, initiates pgRNA encapsidation and replication; B: In vitro priming assay. P protein with its Terminal protein (TP), reverse transcriptase (RT), and RNase H (RH) domains can be activated by reticulocyte lysate (RL), or individual factors (X, Y, Z), to bind ε; ε may be supplied as small RNA covering just the hairpin structure. Upon addition of α32P-dNTPs, P uses ε as template to copy 3 to 4 nt from the ε bulge region; by the covalent linkage of the 5´ nt to a tyrosine residue in TP, P protein becomes radioactively labeled, providing a sensitive assay for activity. In vitro priming does thus far not work with human HBV P protein and ε RNA.
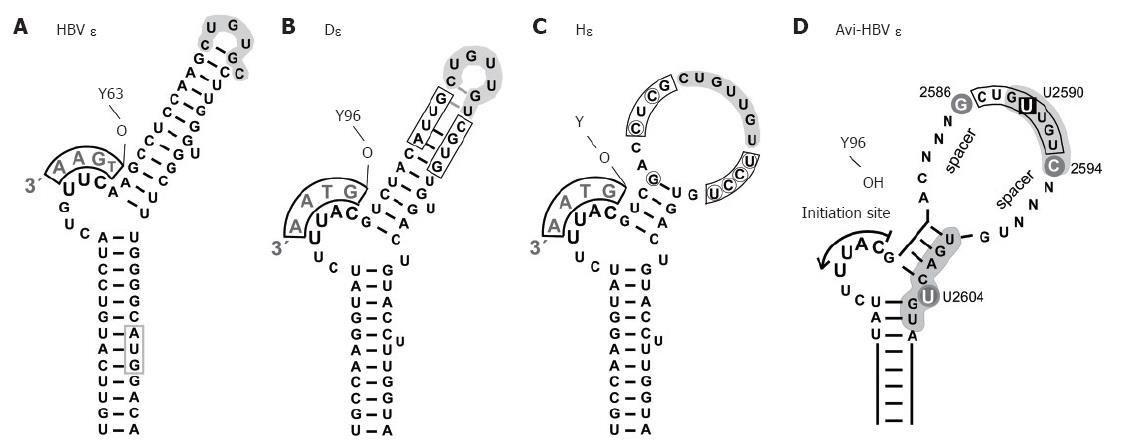
Figure 4 Secondary structures of hepadnaviral ε signals.
A: HBV ε. The entire hairpin, including in the upper stem region, is stably base-paired; formation of a stable tri-loop, as indicated, is confirmed by direct NMR analysis[68]. The conventional annotation for the loop sequence is high-lighted by grey shading. The bulge-templated DNA oligonucleotide and the priming Y63 are indicated; B: DHBV ε. The overall 2D structure is similar to that of HBV ε, including a largely base-paired upper stem[69], as confirmed by preliminary NMR data. The boxed positions were randomized in a SELEX approach, and P binding individuals were selected from the corresponding RNA library[71]; C: HHBV ε (Hε). Hε shows substantial sequence variation to D: (encircled nt), leading to a largely open upper stem structure; D: Generalized secondary structure of an avihepadnaviral ε signal. The scheme summarizes major determinants for productive interaction with P protein[71]. Grey background indicates nt positions that are probably in contact with protein. The bulge and its immediate vicinity, particularly the tip of the lower stem, are essential for P binding whereas the loop appears critically involved in the transition to a productive initiation complex[109]. U2590 > C or U2604 > G mutations abrogate, or strongly reduce, P binding whereas G2586 and C2594 do not affect binding but are important for priming. The major role of the nt termed N may be to provide a proper spacing to the bulge element; their sequence is not important as long as formation of highly stable base-pairing is prevented.
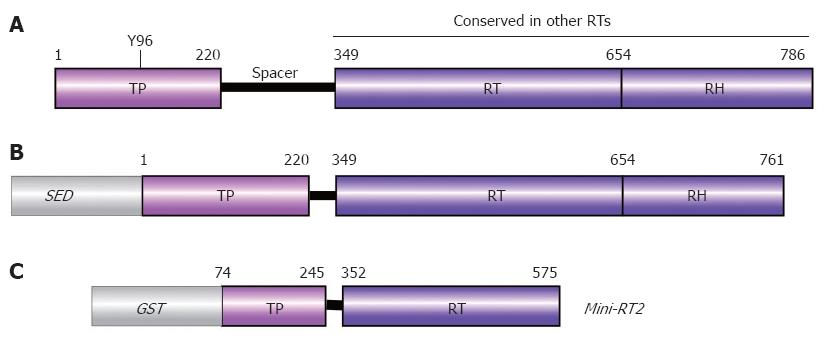
Figure 5 Domain structure of P protein.
A: Authentic DHBV P protein. Numbers are aa positions for DHBV P protein. The priming Tyr residue Y96 is indicated; B: Typical recombinant P protein construct. For solubility, a heterologous solubility enhancing domain (SED) such as NusA, GrpE, or GST is required, and a short stretch of C terminal aa must be removed. Deletion of the spacer has no negative effects on in vitro activity; C: Mini-RT2. This heavily truncated recombinant DHBV P protein requires mild detergent, but no chaperones for priming activity.
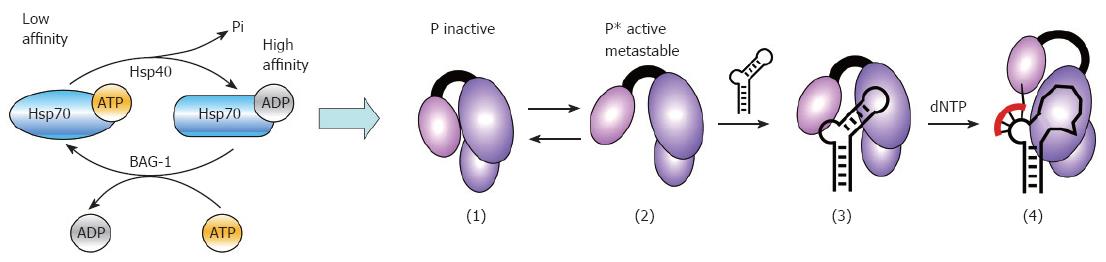
Figure 6 Model for Hsp40/Hsp70 mediated in vitro activation of P protein.
The low ATPase activity of Hsp70 is stimulated by Hsp40, yielding high substrate affinity Hsp70/ADP. A new cycle of substrate release and folding requires exchange of the ADP for new ATP which is stimulated by nucleotide exchange factors such as BAG-1. This ATP-dependent Hsp70 cycling applies to the chaperone´s global folding activities but likely also to P activation: in the inactive state of P (1), the ε RNA binding pocket is inaccessible; Hsp40/Hsp70 activation creates active P* (2) which is able to bind ε RNA (3); P* is metastable, and decays to the inactive state (1) within minutes. Maintaining a steady-state level of P* requires constant re-activation by Hsp40/Hsp70 and thus a continuous supply of fresh ATP. Complexes containing priming-competent ε RNAs (3) undergo induced-fit rearrangements in the RNA and the protein, enabling them to initiate DNA synthesis upon dNTP addition (4). Several Dε variants bind P but do not act as template; most likely they are trapped at stage (3). The same may hold for human HBV P protein complexes with HBV ε RNA.
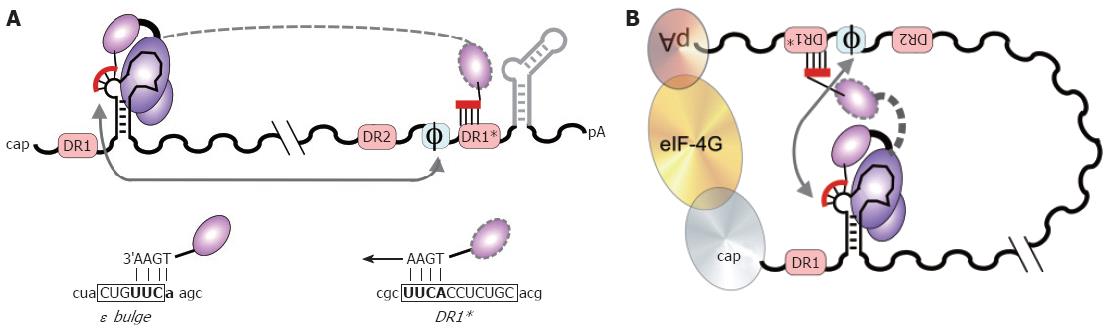
Figure 7 DNA primer translocation (first template switch).
P copies 3 to 4 nt from the 5´ ε bulge, yielding the TP-linked DNA oligonucleotide which is translocated to the complementary motif in the 3´ proximal DR1*. A: Linear representation. DR1* is nearly 3 kb apart from 5´ ε in primary sequence, and numerous other UUCA motifs are not used as acceptors. Φ denotes a newly identified cis-element with partial sequence complementarity to the 5´ half of ε. 3´ ε (light grey) is dispensable. The HBV specific sequences in the ε bulge, and in DR1* are shown below in capitals, flanking sequences in lower case; B: Models for juxtaposition of 5´ ε and DR1*. A general mechanism would be closed loop formation[80] of the pgRNA by cap-binding and poly-A binding factors (ovals), e.g. via elongation initiation factor 4G (eIF-4G; large oval). More specifically, ε might base-pair with Φ[146,147], as indicated by the grey arrows. In such an arrangement, a small movement, rather than a big jump, of TP with the bound DNA primer (dashed outline) would suffice for specific translocation to DR1*.
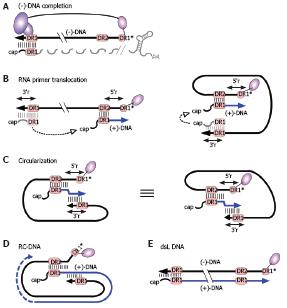
Figure 8 RC-DNA formation.
A: (-)-DNA completion. The DNA primer, still linked to TP, is extended from DR1* to the 5´ end of pgRNA. The RNA is simultaneously degraded by the RH domain, except for its capped 5´ terminal region including 5´ DR1; the fate of the poly-adenylated 3´ end is unclear; B: RNA primer translocation (second template switch). The RNA primer translocates to DR2, and is extended to the 5´ end of (-)-DNA. 3´ r and 5´ r denote an about 10 nt redundancy on the (-)-DNA. As above, several cis-elements appear to promote close proximity of the DR1 donor and the DR2 acceptor, as schematically indicated in the right hand figure; C: Circularization (third template switch). Having copied 5´ r, the growing 3´ end of the (+)-DNA switches to 3´ r on the (-)-DNA, enabling further elongation. This reaction must involve juxtaposition of 5´ r and 3´ r. For easier comprehension, the switch is also depicted on the basis of the representation shown on the right of Figure 8B; both are topologically equivalent; D: RC-DNA. Extension on the (-)-DNA template creates a set of (+)-DNA strands of various length; E: Double-stranded linear (dsL) DNA. This minor DNA form originates when the RNA primer, having failed to translocate to DR2, is extended from its original position ("in situ priming").
- Citation: Beck J, Nassal M. Hepatitis B virus replication. World J Gastroenterol 2007; 13(1): 48-64
- URL: https://www.wjgnet.com/1007-9327/full/v13/i1/48.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i1.48