©2006 Baishideng Publishing Group Co.
World J Gastroenterol. Feb 28, 2006; 12(8): 1287-1291
Published online Feb 28, 2006. doi: 10.3748/wjg.v12.i8.1287
Published online Feb 28, 2006. doi: 10.3748/wjg.v12.i8.1287
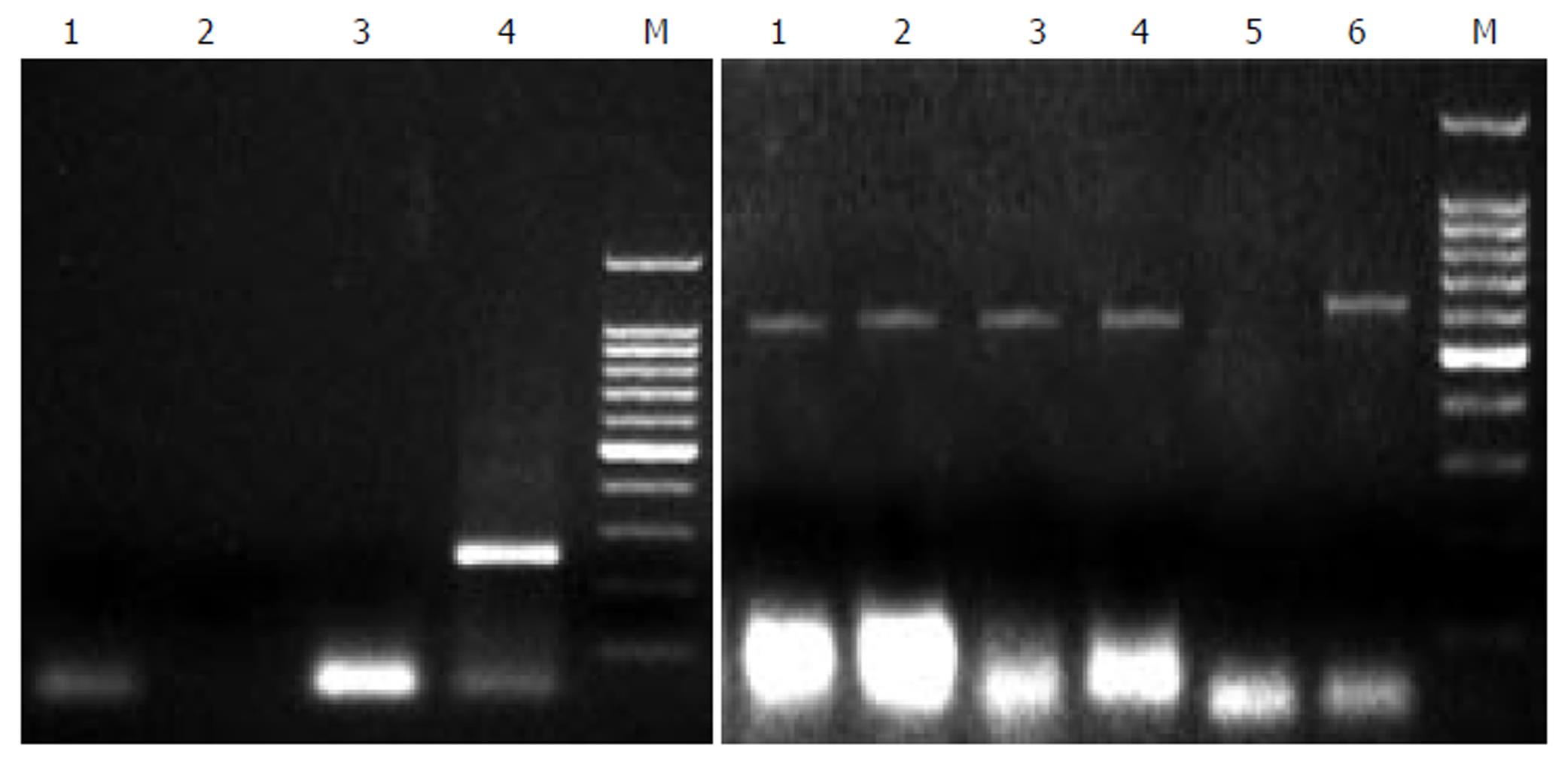
Figure 1 PCR analysis of PERV proviral DNA in different liver tissues.
Left: 1: normal person liver tissue, 2: HBV positive liver tissue, 3: HCV positive liver tissue, 4: second PCR of pig liver tissue, 5: Marker. Right: 1, 2, 3, 4, 6: pig liver cells, 5: nagative control.
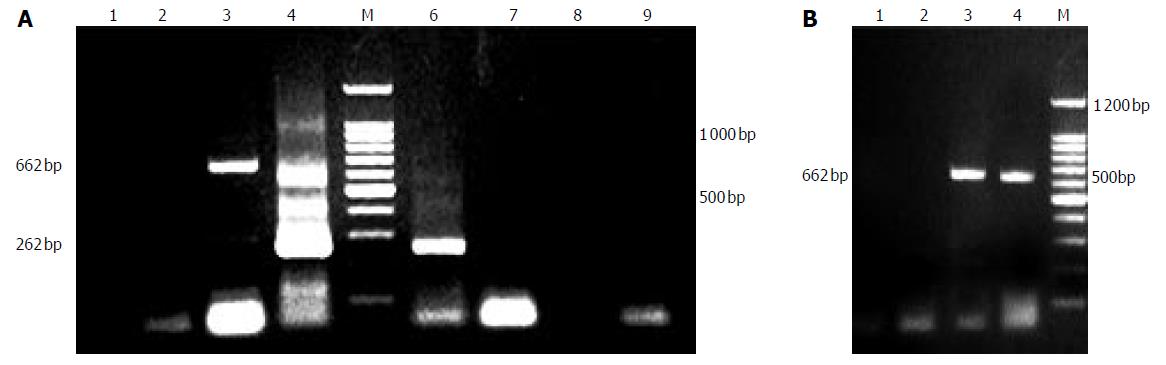
Figure 2 A: PERV RNA detection of supernate.
1:supernate of d 1, 2: supernate of d 3; 3:no RT PCR of d 3; 4:second PCR of d 3; 6: second PCR of d 5; 7: RT-PCR of d 7; 8: RT-PCR of d 9; 9: no RT PCR of d 9; B: PERV permeation of bioreactor. 1: intraluminal supernate of d 3 (no RT PCR), 2: extraluminal supernate of d 3 (no RT PCR), 3: intraluminal supernate of d 3 (RT-PCR); 4: extraluminal supernate of d 3 (RT-PCR).
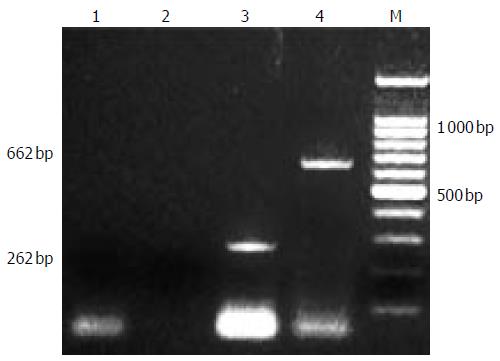
Figure 3 infection of PERV.
1,2 RT-PCR and second PCR of cultured fetal hepatocytes, 3,4: positive control.
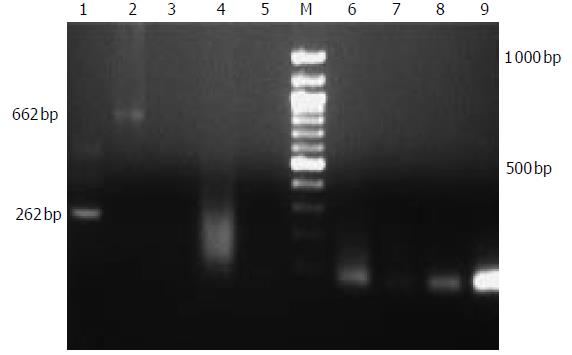
Figure 4 PERV detection of patients treated with EBLSS.
1:second PCR of positive control, 2: positive control, 3:control; 4-9:RT-PCR and second PCR of patients.
- Citation: Wang HH, Wang YJ, Liu HL, Liu J, Huang YP, Guo HT, Wang YM. Detection of PERV by polymerase chain reaction and its safety in bioartificial liver support system. World J Gastroenterol 2006; 12(8): 1287-1291
- URL: https://www.wjgnet.com/1007-9327/full/v12/i8/1287.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i8.1287