©2006 Baishideng Publishing Group Co.
World J Gastroenterol. Dec 28, 2006; 12(48): 7852-7858
Published online Dec 28, 2006. doi: 10.3748/wjg.v12.i48.7852
Published online Dec 28, 2006. doi: 10.3748/wjg.v12.i48.7852
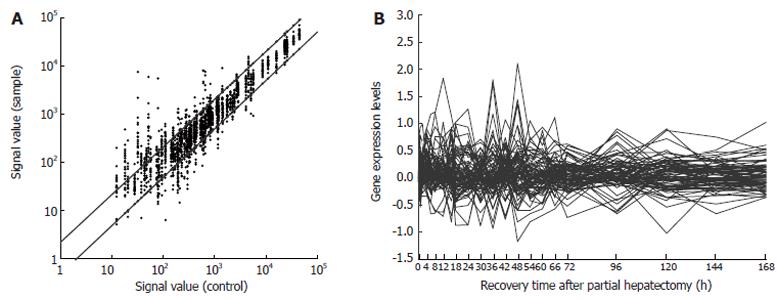
Figure 1 Expression frequency (A) and changes (B) of 85 innate immune response-associated genes during rat liver regeneration.
Data detected by rat genome 230 2.0 array were analyzed and graphed by Microsoft Excel. The dots above bias indicate that the expression of genes was increased more than two folds and up-regulated 350 times, the dots under bias indicate that the expression of genes was decreased more than two folds and down-regulated 129 times, the dots between biases indicate that the expression of genes has no alteration. The expression of 59 genes was increased 2-128 folds, while the expression of 44 genes was increased 2-10 folds.
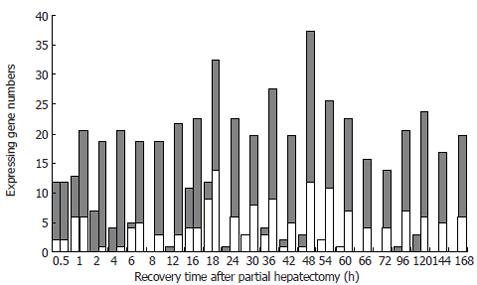
Figure 2 Initial and total expression profiles of 85 innate immune response-associated genes at each time point of liver regeneration.
Grey bars: up-regulated expression gene; white bars: down-regulated expression gene; blank bars: initially expressed genes in which up-regulated genes are predominant in the forepart, and down-regulated genes in the prophase and metaphase, whereas very few in the anaphase; dotted bars: the total number of expressed genes, in which the expression of some genes is up-regulated and the expression of others is down-regulated during LR.
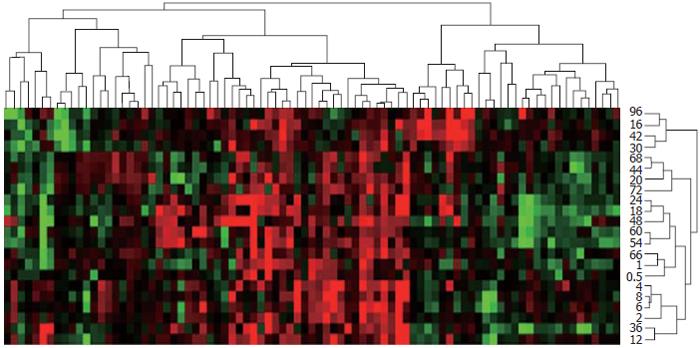
Figure 3 Expression similarity and time relevance clusters of 85 innate immune response-associated genes during liver regeneration.
Data detected by rat genome 230 2.0 array were analyzed by H-clustering. Red indicates up-regulated gene expression chiefly associated with cellular immunity; green indicates down-regulated gene expression mainly associated with molecular immunity; black indicates meaningless change in gene expression. The upper and right trees show the expression similarity and time series clusters, by which the above genes were classified into 5 and 14 groups respectively.
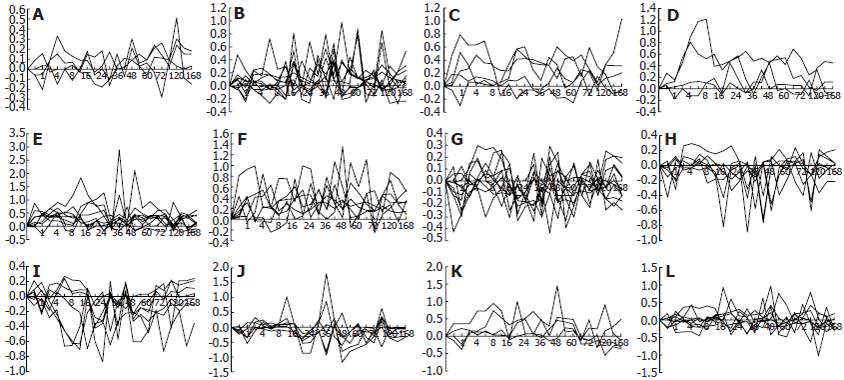
Figure 4 Expression patterns of 85 innate immune response-associated genes during liver regeneration.
Twenty-eight expression patterns were obtained by the analysis of data detected by rat genome 230 2.0 array with Microsoft Excel. A-F: 41 up-regulated genes; G-I: 26 down-regulated genes; J-L: 18 up/down-regulated genes. X-axis represents recovery time after PH (h); Y-axis shows logarithm ratio of the signal values of genes at each time point to control.
- Citation: Chen GW, Zhang MZ, Zhao LF, Xu CS. Expression patterns and action analysis of genes associated with physiological responses during rat liver regeneration: Innate immune response. World J Gastroenterol 2006; 12(48): 7852-7858
- URL: https://www.wjgnet.com/1007-9327/full/v12/i48/7852.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i48.7852