©The Author(s) 2004.
World J Gastroenterol. Nov 1, 2004; 10(21): 3132-3136
Published online Nov 1, 2004. doi: 10.3748/wjg.v10.i21.3132
Published online Nov 1, 2004. doi: 10.3748/wjg.v10.i21.3132
Figure 1 Genotype-specific sites recognized by restriction enzymes.
GeneBank accession numbers and consensus sequences from HBV genotypes A to H are listed. The shaded letters indicate the sequences recognized by the relevant enzyme. StyI recognizes sequence C|C (A/T) (A/T) GG; BsrI, C|CAGT; DpnI, GA|TC; HpaII, C|CGG; EaeI, (C/T) |GGCC (A/G). *nt position number is from the unique EcoRI site according to the reference sequence × 001587 (genotype C).
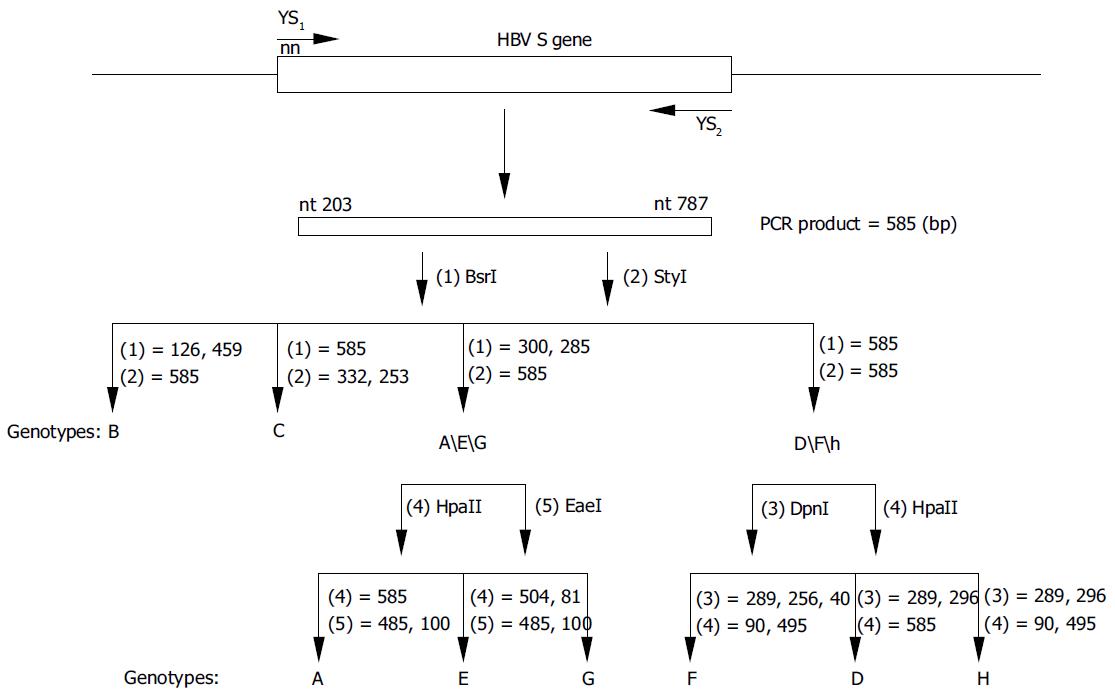
Figure 2 Diagrammatic representation of the position of PCR primers and RFLP analyses for HBV genotyping.
The second round PCR products, which were 585 bp, were digested by (1) BsrI, (2) StyI, (3) DpnI, (4) HpaII and (5) EaeI. Genotypes B and C could be typed in one step using parallel digestion with BsrI and StyI.
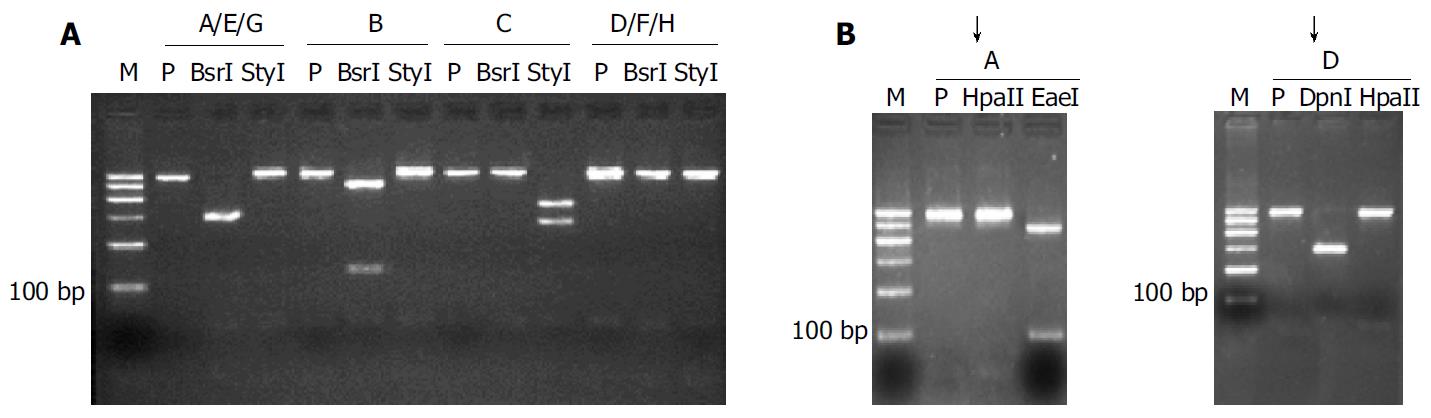
Figure 3 RFLP of S amplicon of HBV from 187 nested PCR-positive serum samples from Chinese carriers.
Lane M: 100 bp ladder from 100-600 bp fragments, P: Undigested PCR product. A: After the first step of parallel digestion with BsrI and StyI, the samples were divided into four groups: Genotypes B, C, A/E/G and D/F/H; B: Groups A/E/G and D/F/H could be further resolved by the second parallel digestion. In this study, genotypes E, F, G and H were not observed, while genotypes B and C were the major ones.
-
Citation: Zeng GB, Wen SJ, Wang ZH, Yan L, Sun J, Hou JL. A novel hepatitis B virus genotyping system by using restriction fragment length polymorphism patterns of
S gene amplicons. World J Gastroenterol 2004; 10(21): 3132-3136 - URL: https://www.wjgnet.com/1007-9327/full/v10/i21/3132.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i21.3132