INTRODUCTION
Artificial intelligence (AI) has emerged as a new tool with a wide applicability and has transformed every aspect of society including medicine. This technology is an assimilation of human intelligence through computer algorithms to perform specific tasks[1-3]. Machine learning (ML) and deep learning (DL) are techniques of AI. A ML system refers to automatically built mathematical algorithms from data sets that form decisions with or without human supervision[1-3]. A DL system is a subdomain of ML in which AI self-creates algorithms that connects multi-layers of artificial neural networks[1-3].
The recent expansion of research involving AI has shed light on the potential applications in gastrointestinal diseases. Researchers have developed computer aided diagnosis (CAD) systems based on DL to enhance detection and characterization of lesions. CAD systems are now being investigated in numerous studies involving Barrett’s esophagus, esophageal cancers, inflammatory bowel disease, and detection and characterization of colonic polyps[4].
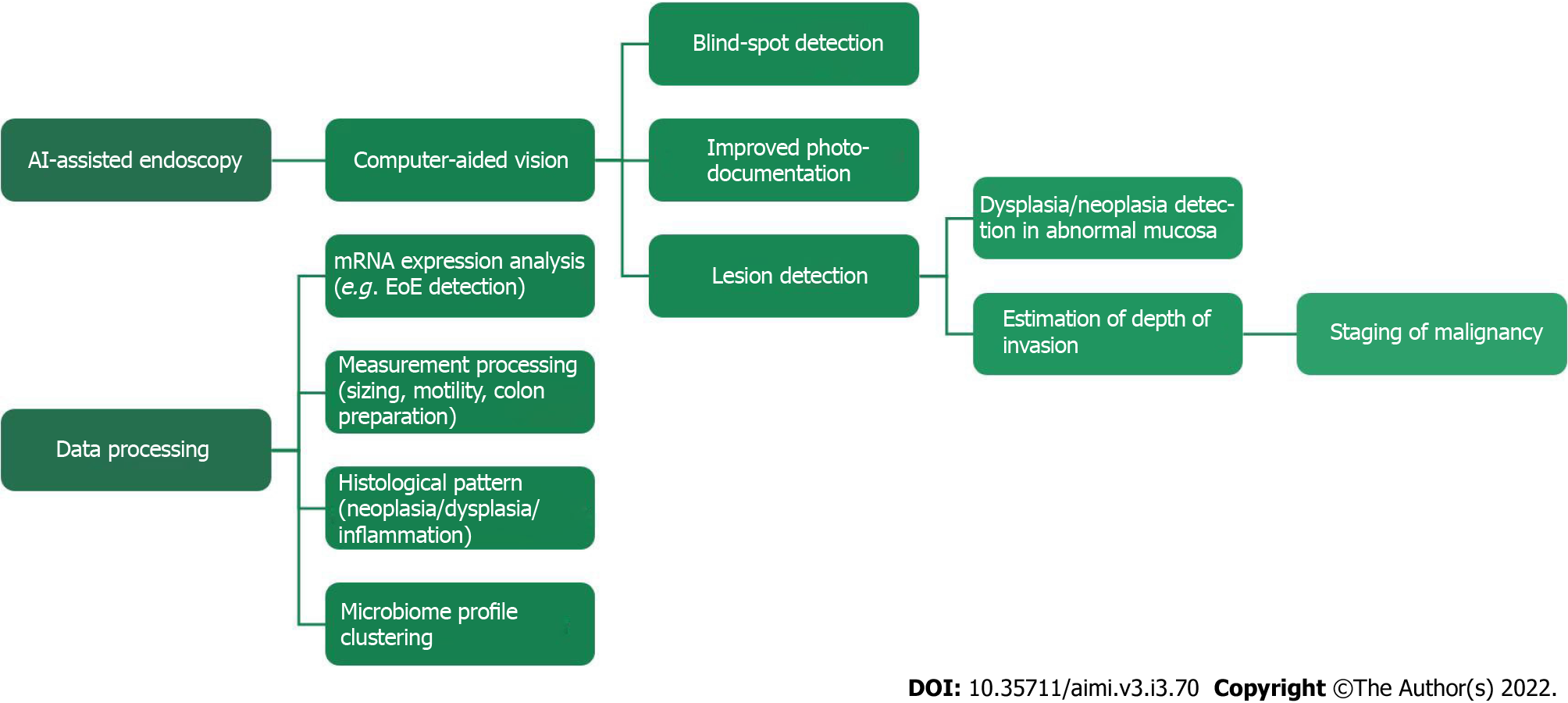
In this review, we aim to evaluate the evidence on the role of AI in endoscopic screening and surveillance of gastric and esophageal diseases. In addition, we also provide the current limitations and future directions associated with eosinophilic esophagitis and esophageal microbiome (Figure 1).
Figure 1 Artificial intelligence -assisted endoscopy and data processing are the currently demonstrated uses for Artificial intelligence.
AI: Artificial intelligence; EoE: Eosinophilic esophagitis; mRNA: Messenger ribonucleic acid.
MATERIALS AND METHODS
A literature search to identify all relevant articles on the use of AI in endoscopic screening and surveillance of gastric and esophageal diseases was conducted. The search was conducted utilizing PubMed, Medline, and Reference Citation Analysis (RCA) electronic database. We performed a systematic search from January 1998 to January 2022 with search words and key terms including “artificial intelligence”, “deep learning”, “neural network”, “endoscopy”, “endoscopic screening”, “gastric disease”, esophageal disease”, “gastric cancer”, “gastric polyps”, “Barrett’s esophagus”, “eosinophilic esophagitis”, “microbiome”.
AI AND GASTRIC POLYPS
Gastric polyps represent abnormal tissue growth, the majority of which do not cause symptoms and, as such, are often found incidentally in patients undergoing upper gastrointestinal endoscopy for an unrelated condition[5]. The incidence of gastric polyps ranges from 1% to 6%, depending on geographical location and predisposing factors, such as Helicobacter pylori (H. pylori) infection and PPI use[6]. While most polyps are not neoplastic, certain subtypes carry malignant potential with a rater of cancerization as high as 20%[7]. Therefore, the primary utility of polyp detection is cancer prevention. The necessity for detection and recognition of precancerous gastric polyps and the fact that most are incidental findings are a crossroad that has helped propel research and advancement in the field of AI computer-assisted systems for upper-endoscopy.
Detection of gastric polyps
One way to increase accurate detection of gastric polyps is by ensuring complete mapping of the stomach during esophagogastroduodenoscopy (EGD). WISENSE is a real-time quality improvement system that uses deep convolutional neural network (DCNN) and deep reinforcement learning to monitor blind spots, track procedural time and, generate photo documentation during EGD. One of the datasets used to train the network of learning and classifying gastric sites utilized 34513 qualified EGD images. Images were labeled into 26 different sites based on the guidelines of the ESGE and Japanese systematic screening protocol. The system was tested using a single-center randomized-control trial. A total of 324 patients were randomized, with 153 of them undergoing EGD with WISENSE assistance. The rate of blind spots (number of unobserved sites in each patient/26) was significantly lower for WISENSE group compared to the control group, 5.86% vs 22.46%. Additionally, the system led to increased inspection time and completeness of photodocumentation[8].
A year after the previously mentioned study, the developers renamed WISENSE to ENDOANGEL and further explored the systems capability of identifying blind spots in three different types of EGD; sedated conventional EGD (C-EGD), non-sedated ultrathin transoral endoscopy (U-toe), and non-sedated C-EGD[9]. ENDOANGEL was tested using a prospective single-center, single-blind, randomized, 3-parallel group study. The study results indicated that with the assistance of ENDOANGEL the blind spot rate was significantly reduced for all three EGD modalities. The greatest reduction was seen in the sedated C-EGD group and demonstrated 84.77% reduction. Non-sedated U-TOE and C-EGD blind spot rate decreased by 24.24% and 26.45%, respectively[9]. The major benefit of ENDOANGEL is that it provided real-time prompting when blind spots were identified, thereby allowing the endoscopist to re-examine the missing parts and improve overall visualization. Furthermore, through reduction in total blind spots the authors extrapolate that ENDOANGEL has the potential to mitigate the skill variation between endoscopists[9].
While neither of the above-mentioned systems are specifically designed for the detection of polyps, these encourage and assist endoscopists in completing complete and thorough visualization of stomach during upper endoscopy, a task that has become more daunting over the years as the workload of endoscopists continues to increase. Multiple research groups have created various automated computer-aided vision methods to help detect gastric polyps in real time. Billah et al[10] proposed a system that uses multiresolution analysis of color textural features. These color wavelet (CW) features are used in conjunction with CNN features of real time videoframes to train a linear support vector machine (SVM). The fusion of all three features then allows the SVM to differentiate between polyp and non-polyp. The program was trained using more than 100 videos from various sources, resulting in greater than 14000 images being used. This proposed model was then tested on a standard public database and achieved a detection rate of 98.65 %, sensitivity of 98.79%, and specificity 98.52%.
One of the commonly encountered problems with regard to developing computer-aided polyp detection systems is identification of small polyps. To address this problem, Zhang et al[11] constructed a CNN using enhanced single shot multibox detector (SSD) architecture that they termed SSD for gastric-polyps (SSD-GPNet). This system was designed to circumvent the problem of lost information that occurs during the process of max-pooling utilized by the SSD feature pyramid during object detection. By reusing this lost information, their new algorithm maximized the quantity of information that could be utilized and therefore increased detection accuracy. The system was tested on 404 images containing gastric polyps, the majority of which were categorized as small. According to the authors, the system was able to achieve real-time gastric polyp detection with a mean average precision of 90.4% utilizing a speed of 50 frames per second[11].
Recently, Cao et al[7] developed a system that further improves upon the traditional feature pyramid to identify small polyps as well as those that are more difficult to distinguish from surrounding mucosa due to similarity in features. Their proposed system contains a ‘feature fusion and extraction module’ which allows the program to combine features from multiple levels of view without diluting the information obtained from adjacent levels. In doing so, program continues to create new feature pyramids which deepens the network, retains more high-level semantic and low-level detailed texture information. The retention and fusion of such information allows the system to distinguish gastric polyps from gastric folds. The system was trained using 1941 images with polyps. To overcome the small data set, the authors utilized random data augmentation which consists of changing image hue and saturation, rotation of the image, etc. The system demonstrated a mean sensitivity of 91.6% and recall of 86.2% (proportion correctly identified true positives), after 10-fold validation testing[7]. Unfortunately, the authors do not provide detection results regarding those polyps they deemed difficult to discern from gastric folds. Nonetheless, the development of an augmented data set and a high level of sensitivity show promise with regards to overall polyp detection rates.
Characterization of gastric polyps
There are numerous types of gastric polyps and most of them do not carry any malignant potential. The two classes of polyps with the highest potential for malignancy are hyperplastic polyps and gastric adenomas. Gastric adenomas, or raised intraepithelial neoplasia, represent direct precursor lesions to adenocarcinoma and rarely appear in the presence of normal gastric mucosa. Instead, they are often found on a background of chronic mucosal injury, such as chronic gastritis and gastric atrophy[6]. Therefore, many of the AI systems that have been developed to assist endoscopists in the prevention of gastric cancer focus on the characterization and identification of known gastric cancer precursor lesions such as gastric atrophy and intestinal metaplasia, rather than characterizing all the various types of polyps. Characterization of gastric polyps relies heavily on image-enhanced endoscopy (IEE). Especially modalities such as narrow-band imaging (NBI) and blue laser imaging with or without magnification.
Xu et al[12] utilized various IEE images to train their DCNN system, named ENDOANGEL, to detect and diagnose gastric precancerous conditions, specifically gastric atrophy and intestinal metaplasia, in real time. The performance of their AI model tested using a prospective video set achieved an accuracy of 87.8%, sensitivity of 96.7% and specificity of 73.0% with regards to identification of gastric atrophy. In the prospective video set test for intestinal metaplasia the system achieved an accuracy, sensitivity, and specificity of 89.8%, 94.6%, and 83.7%, respectively[12]. Additionally, the system performance was tested against that of endoscopist with varying degrees of expertise (for a subset 24 patients). Overall, the program performed similarly to 4 expert endoscopists (those with 5 or more years of training including 3 or more in IEE). Compared to 5 nonexpert endoscopists (those with 2 years of endoscopic experience and 1 year of experience in IEE) who had a mean accuracy of 75.0%, sensitivity of 82.8% and specificity of 59.4% for GA and an accuracy of 73.6%, sensitivity of 73.8%, and specificity of 73.3% for IM, ENDOANGEL performed significantly better[12].
Limitations of AI in gastric polyps
To the best of our knowledge, there have been no randomized control trials to evaluate the clinical efficacy of AI automated gastric polyp detection systems. However, the accuracy, sensitivity, and specificity of those mentioned here, as well as others not mentioned, indicate great potential in assisting endoscopist to detect gastric polyps. With the further development of AI systems to not only detect but, to characterize these gastric lesions, the potential clinical utility is further increased. AI systems with fully developed CADe and CADx can be developed to aid rapid and effective decision making for identifying lesions that should be targeted for biopsy. Such systems may also improve other patient outcomes by mitigating the difference in endoscopist experience.
AI AND GASTRIC CANCER
Gastric cancer (GC) is the fifth most common cancer in the world and the fourth most fatal cancer[11]. The 5-year survival rate is greater than 90% when diagnosed at early stages, making early detection particularly important[7]. Alarmingly, in 2019, more than 80% of GCs in China were diagnosed at advanced stages, signifying inadequate early detection[12]. Risk factors for GC include H. pylori infection, alcohol use, smoking, diet, race and gender[13]. Due to the non-specific nature of symptoms, most GC is usually diagnosed at later stages which makes prognosis poor[14].
Although endoscopic imaging is the most effective method of detection, visualization can be difficult. The reasons for this include the subtle changes in mucosa (elevations, depressions, redness or atrophy) that can be mistaken for gastritis or intestinal metaplasia, especially when found in a region with background gastritis[15]. Further, the subjective nature of identification makes detection endoscopist dependent with reported miss rates as high as 14% and 26%[15,16]. In addition to the limitations in detecting mucosal changes, endoscopy is historically poor at predicting depth of invasion with studies reporting only 69% to 79% accuracy[17]. This is important because accurately predicting depth of invasion can aid in guiding management and surgical planning.
Over the past several decades, AI has expanded towards new horizons in medicine and image recognition. Recently, DL has become more widely applied in the prevention and detection of GC. Medical image recognition in locating tumors is called “image segmentation”. Importantly, image segmentation determines diagnostic accuracy for evaluation and surgical planning in GC. DL has been shown to improve image segmentation via three networks; supervised network, semi-supervised network, and unsupervised network[18]. Supervised learning networks comprise the majority. These networks use large data sets that are preemptively labeled. Convolutional Neural Networks (CNN) are supervised learning networks which have demonstrated high performance in image recognition tasks[18].
Prevention, detection and classification of gastric cancer
For prevention of GC, it is important to optimize the diagnosis and eradication of H. pylori. In 2018, Itoh et al[19] developed a CNN-based system which was trained on 149 images to diagnose H. pylori. The results showed 86.7% sensitivity and 86.7% specificity which significantly outcompetes traditional endoscopy and the researchers concluded that CNN-aided endoscopy may improve diagnostic yield in H. pylori endoscopy.
A 2020 systematic review and meta-analysis. reviewed 8 studies with 1719 patients and found a pooled sensitivity and specificity of 0.87 (95%CI 0.72-0.94) and 0.86 (95%CI 0.77-0.92), respectively in predicting H. pylori infection. In addition, the study showed an 82% accuracy of AI for differentiating between post eradication images and non-infected images[20]. The authors were also able to identify 2 studies where discrimination using AI, between H. pylori infected and post-eradicated images was analyzed, revealing an accuracy of 77%. While the authors state external validity as a limitation of this study, the results cannot be ignored in the context of prior studies. Accordingly, AI may have a role in diagnosis as well as confirmation of treatment.
Along with eradication of H. pylori, prevention also comes in the form of detecting precancerous lesions. These lesions include erosion, polyps and ulcers which may develop into gastric cancer if they are not detected early. In 2017, Zhang et al[21] developed a CNN known as the Gastric Precancerous Disease Network (GPDNET) to categorize precancerous gastric disease. This AI demonstrated an accuracy of 88.90% in classifying lesions as either polyps, erosions or ulcers.
As previously mentioned, GC is often discovered in late stages, which thereby makes improvements in early detection, particularly important. Deep learning algorithms have shown promise with this regard. A study by Li et al[22] demonstrated significantly higher diagnostic accuracy in CNN trained (90.91%) endoscopy compared to non-experts (69.79 and 73.61%) (P < 0.001 with kappa scores of 0.466 and 0.331). The researchers looked at CNN-based analysis of gastric lesions observed by magnifying endoscopy with narrow band imaging (M-NBI) and found a 91.8% sensitivity, 90.64 specificity and 90.91 accuracy in diagnosing early gastric cancer (EGC). While specificity was like that of experts, sensitivity of EGC detection was superior to both experts (78.24 and 81.18) and non-experts (77.65 and 74.12). The researchers attributed this to a lack of subjectivity which is inherent to human endoscopy. Ikenoyama et al[23] constructed their CNN using 13584 images from 2639 early GC lesions and compared its diagnostic ability to 67 endoscopists. Results showed faster processing as well as a 26.5% higher diagnostic sensitivity in CNN compared to endoscopists. This further demonstrates the potential for AI to improve efficiency in diagnosing GC.
The role of AI is not limited to early detection. Hirasawa et al[24] constructed a CNN trained with 13584 images to detect both early (T1) and advanced GC (T2-4). They demonstrated an overall sensitivity of 92.2% in diagnosing gastric cancer. The diagnostic yield was further accentuated at diameters of 6mm or greater with a sensitivity of 98.6%. All invasive lesions were correctly identified as cancer during this study. Despite these promising results, there were false positives that lead to a positive predictive value (PPV) of only 30.6%.
In addition to CNN, fully convolutional neural networks (FCN) use pixel level classification to allow for more robust image segmentation[25]. When it comes to distinguishing cancer from precancerous disease, FCN has shown promise. In 2019, Lee et al[26] used data from 200 normal, 220 ulcer and 367 cancer cases to build the Inception-ResNet-v2 FCN which was able to distinguish between cancer and normal as well as cancer and ulcer at accuracies above 90%. In a 2019 study by Nguyen et al Inception-ResNet-v2 was used to further classify neoplasms based on severity. Five categories were assessed: EGC, advanced GC, high grade dysplasia, low grade dysplasia and non-neoplasm. The result was a weighted average accuracy of 84.6% in classifying neoplasm[27].
Depth of invasion of gastric cancer
Depth of invasion is an important characteristic when it comes to accordant direction for best management of GC[17]. The current evidence suggests that early stages of EGCs with depth limited to the mucosal (M) or superficial submucosal layers (SM1) can be managed with endoscopic submucosal dissection or endoscopic mucosal resection[17]. Invasion into the deeper submucosal layer will require surgery. In 2018, Zhu et al[17] built a CNN computer-aided detection (CNN-CAD) system to determine depth of invasion of GC. The results showed accuracy of 89.16% which was significantly higher than that of endoscopists (69% to 79%). PPV and NPV were 89.66% and 88.97%, respectively. Endoscopists had values of 55.86% and 91.01%. This enhanced ability to predict invasion supports the assertion that CNN has shown utility in helping endoscopists detect, classify, and predict prognosis of GC.
Limitations of AI in gastric cancer
Supervised learning networks show promise in the prevention of cancer through detection of H. pylori and precancerous lesions as well as promise in detection and classification of neoplasm. AI has not only demonstrated superiority to traditional endoscopists when it comes to identifying GC stage but also at determining depth of invasion which can dramatically improve prognosis in a disease with inadequacy of early detection. There is utility when it comes to helping less experienced endoscopists. Despite their superior diagnostic efficacy, supervised learning networks are not immune to false positives and false negatives. Because they rely heavily on the quality and quantity of learning samples, they may interpret poor images of intestinal metaplasia or atrophy as GC and are data dependent[25]. Semi-supervised and unsupervised learning networks are potential alternatives as they are not entirely data dependent[18].
AI AND BARRETT’S ESOPHAGUS
The American Cancer Society’s estimates about 19260 new cases of esophageal cancer (EC) diagnosed (15310 in men and 3950 in women) and about 15530 deaths from EC (12410 in men and 3120 in women) in the United States in 2021[28]. It is the seventh most common cancer and the sixth leading cause of cancer related mortality worldwide[29]. The two major histological types of EC are adenocarcinoma (AC) and squamous cell carcinoma (SCC)[30]. For SCC alone, the primary causal risk factors vary geographically. Over the past 40 years, the incidence of AC, which typically arises in the lower third of the esophagus, has risen faster than any other cancer in the Western world, and rates continue to rise even among new birth cohorts. Conversely, the incidence of SCC has declined in these same populations. As such, AC is now the predominant subtype of esophageal cancer in North America, Australia and Europe. Like AC, the incidence of Barrett esophagus has increased in many Western populations[31].
Barret’s esophagus (BE) is a change of the normal squamous epithelium of the distal esophagus to a columnar-lined intestinal metaplasia, and the main risk factors associated with its the development are long-standing gastroesophageal reflux disease (GERD), male gender, central obesity, and age over 50 years[32]. It is thought to follow a linear progression from nondysplastic BE to low-grade dysplasia to high-grade dysplasia and finally to cancer. The presence of regions of dysplasia in BE increases the risk of progression and guides treatment considerations. Early detection of dysplastic lesions and cancer confined to the mucosa allows for minimally invasive curative endoscopic treatment, which provides a less invasive method of treatment than surgical resection and/or neo adjuvant therapy for advanced lesions. However, the evaluation and assessment of BE is challenging for both expert and nonexpert endoscopists. The appearance of dysplasia may be subtle, and segmental biopsy samples may not detect patchy dysplasia[33,34].
Current challenges in Barrett’s esophagus
Results from a multicentric cohort study support that missed esophageal cancer is relatively frequent at routine upper gastrointestinal endoscopies in tertiary referral centers, with an overall MEC rate as high as 6.4% among newly diagnosed esophageal cancer patients[35]. Additionally, a recent meta-analysis showed a high miss rate of 25% for high grade dysplasia and cancer within 1 year of a negative index examination, the reasons for this are likely multifactorial, including the lack of recognition of subtle lesions, lack of detailed inspection of the esophageal mucosa, non-optimum cleaning techniques, and less experienced endoscopists[34].
Optical identification and diagnosis of dysplasia would guide treatment decisions during endoscopy for BE. The limitations of current screening and surveillance strategies impulse to improve diagnostic accuracy and risk stratification of patients with BE. In recent years, many new endoscopic techniques have been developed, such as magnification endoscopy, chromoendoscopy, confocal laser endomicroscopy, and volumetric laser endomicroscopy, most of which are expensive and take a long time for endoscopists to learn. Differences in endoscopists' interpretations of the images can also lead to differences in diagnosis[36].
AI and convolutional neural network
A proposed use of AI during upper endoscopy will be with live video images that will be sent to the AI application and analyzed in real time. The application will be able to detect areas suspicious for neoplasia and measure the size and morphology of lesions. It will alert the endoscopist to suspicious areas either with a screen alert or location box. The endoscopist can then decide if the area needs to be sampled based on the characterization provided by the machine or managed endoscopically[34]. Therefore, AI can assist in by using methods of DL to identify and process in real-time endoscopic data that may not consciously appreciated by humans such as subtle changes in color and texture to aid in taking targeted biopsies rather than random biopsies.
AI uses several machine learning methods, one that is frequently used is CNN, a form of DL which receives input (e.g. endoscopic images), learns specific features (e.g. pit pattern), and processes this information through multilayered neural networks to produce an output (e.g. presence or absence of neoplasia). Several layers of neurons can exist to make a single decision to call a grouping of pixels on an image either normal tissue or dysplasia. The advantages that AI appears to confer per-endoscopy is a removal of the inter-observer or intra-observer variability in identification of non-normal lesions, combined with rapid, objective analysis of all visual inputs in such a way that is consistent and not subject to fatigue. This advanced technology of CAD can allow endoscopists to take targeted, high-yield biopsies in real-time. Compared to taking random biopsies per the Seattle protocol or using enhanced imaging, CAD may increase efficiency and accuracy for making a diagnosis by limiting the chance of missing neoplastic mucosa. Moreover, CAD may decrease risk by decreasing sedation time secondary to decreased procedure length[37].
AI use with white light imaging
Van der Sommen et al[38] in 2016 collected 100 images from 44 BE patients and created a machine learning algorithm which used texture and color filters to detect early neoplasia in BE. The sensitivity and specificity of the system were 83% for the per-image analysis and 86% and 87% for the per-patient analysis, respectively. Therefore, the automated computer algorithm developed was able to identify early neoplastic lesions with reasonable accuracy, suggesting that automated detection of early neoplasia in Barrett’s esophagus is feasible.
In a study by de Groof et al[39], six experts identified likely neoplastic tissue in the same image and used these expert-delineated images to train the computer algorithm to identify neoplastic BE and non-dysplastic BE in test cases. The resulting sensitivity and specificity of the computer algorithm was 0.95 and 0.85 respectively. de Groof et al[40] developed a deep learning system using high-definition white light endoscopy images of over 10000 images of normal GI tract followed by 690 images of early neoplastic lesions and 557 non dysplastic Barrett’s epithelium to detect, delineate the lesion, and pinpoint high yielding biopsy sites withing the lesion. This group was able to externally validate their CAD system demonstrating a better accuracy of 88% in detecting early neoplastic lesions compared with an accuracy of 73% with endoscopists. Ebigbo et al[41] were also able to validate a CNN system to detect EAC in real time with the endoscopic examination of 14 patients using 62 images and showed a sensitivity of 83.7% and specificity of 100%.
Hashimoto et al[42] collected 916 images from 70 patients with early neoplastic BE and 916 control images from 30 normal BE patients and then trained a CNN algorithm on ImageNet. The researchers analyzed 458 images using the CNN algorithm. The accuracy, sensitivity, and specificity of the system for detecting early neoplastic BE were 95.4%, 96.4%, and 94.2%, respectively.
AI use with volumetric laser endomicroscopy and confocal laser endomicroscopy
The volumetric laser endomicroscopy system has the capacity to provide three-dimensional circumferential data of the entire distal esophagus up to 3-mm tissue depth. This large volume of data in real-time remains difficult for most experts to analyze. AI has the potential to better interpret such complex data[43].
Interpretation of volumetric laser endomicroscopy (VLE) images from BE patients can be quite difficult and requires a steep learning curve. An AI software called intelligent real-time image segmentation has been developed to identify VLE features by different color schemes. A pink color scheme indicates a hyper-reflective surface which implies increased cellular crowding, increased maturation, and a greater nuclear to cytoplasmic ratio. A blue color scheme indicates a hypo-reflective surface which implies abnormal BE epithelial gland morphology. An orange color scheme indicates lack of layered architecture which differentiates squamous epithelium from BE[44].
Swager et al[45], created an algorithm to retrospectively identify early BE neoplasia on ex vivo VLE images showing a sensitivity of 90% and specificity of 93% in detection with better performance than the clinical VLE prediction score. A CAD system reported by Struyvenberg et al[46] analyzed multiple neighboring VLE frames and showed improved neoplasia detection in BE with an area under the curve of 0.91.
Future of AI and applications in Barrett’s esophagus
Ali et al[47] at the University of Oxford reported on one a deep learning tool to automatically estimate the Prague classification and total area affected by columnar metaplasia in patients with Barrett's esophagus. They propose a novel methodology for measuring the risk score automatically, enabling the quantification of the area of Barrett’s epithelium and islands, as well as a 3-dimensional (3D) reconstruction of the esophageal surface, enabling interactive 3D visualization. This pilot study used a depth estimator network is used to predict endoscope camera distance from the gastric folds. By segmenting the area of Barrett’s epithelium and gastroesophageal junction and projecting them to the estimated mm distances, they were able to measure C&M scores including the area of Barrett’s epithelium. The derived endoscopy artificial intelligence system was tested on a purpose-built 3D printed esophagus phantom with varying areas of Barrett’s epithelium and on 194 high-definition videos from 131 patients with C&M values scored by expert endoscopists. The endoscopic phantom video data demonstrated a 97.2% for C&M and island measurements, while the accuracy for the area of Barrett’s epithelium it was 98.4% compared with ground-truth[47].
This is the first study to demonstrate that Barrett’s circumferential and maximal lengths and total affected area can be automatically quantified. While further optimization and extensive validation are required, this tool may be an important component of deep learning-based computer-aided detection systems to improve the effectiveness of surveillance programs for Barrett’s esophagus patients[48].
The studies show promising results and as AI systems develop, it will be important that they are tested and validated in real-world settings, in diverse patient populations, with physicians of varying expertise, with different endoscope types and in different practice settings. Commercially developed AI will need to demonstrate cost-effective care that will provide meaningful value and impact on patient care and outcomes. The field continues to expand and promises to impact the field of BE detection, diagnosis, and endoscopic treatment[33,49].
ACHALASIA AND AI
Achalasia is an esophageal motility disorder characterized by impaired peristalsis and relaxation of the lower esophageal sphincter. While the pathophysiology is incompletely understood, it is thought to be related to loss of inhibitory neurons in the myenteric plexus. Symptoms include dysphagia to both solids and liquids as well as heartburn, chest pain and other nonspecific symptoms. In fact, 27%-42% of patients are initially misdiagnosed as GERD[50].
High-resolution manometry (HRM) is the gold standard[51]. A limitation of manometry is that it cannot differentiate between achalasia and pseudo achalasia, a disorder which is often malignancy presenting as achalasia[52]. As such, the utility of endoscopy comes in ruling out malignancy and endoscopic biopsy is an important part of the diagnostic algorithm. Endoscopy can also be used to rule out other obstructive lesions or GERD[53]. However, HRM is vital in classification of achalasia subtypes which guides treatment and prognosis.
The Chicago Classification system is based on manometric differences between three subtypes. All three have impaired EGJ relaxation[54]. Subtype 1 has aperistalsis with the absence of pan esophageal pressurization. Subtype 2 has aperistalsis with pressurization greater than 30 mmHg and subtype three is characterized by abnormal spastic contractions with or without periods of pan esophageal pressurization. While types 1 and 2 can be corrected with Heller myotomy, type 3 patients are more likely to benefit from more extensive myotomy[55].
Functional lumen imaging probe and AI
The functional lumen imaging probe (FLIP) device that uses high resolution impedance planimetry to measure cross sectional area and pressure to provide a 3D model of achalasia. It has been shown to be just as good as manometry in diagnosing achalasia and has also shown application in cases where clinical suspicion is high, but manometry is equivocal[56]. Because FLIP is performed during endoscopy, it can help identify patients who do not respond to manometry.
Despite its ability to diagnose achalasia, FLIP has limited data available in its ability to differentiate between achalasia subtypes. If it were able to do this, it could essentially combine the steps of endoscopic evaluation, diagnosis, and classification of achalasia. Machine learning may have a role here.
In 2020, Carlson et al[57] were able to demonstrate the application of supervised machine learning in using FLIP to characterize achalasia subtypes in a study of 180 patients. The AI was able to differentiate type 3 achalasia from non-spastic subtypes with an accuracy of 90% while the control group did so with an accuracy of 78%. The machine was also able to further classify achalasia into subtype 1, 2 and 3 with an accuracy of 71% compared to the 55% accuracy of the control group. This is an important application given the differences in prognosis and management based on subtype.
Achalasia and cancer
Esophageal cancer is a rare consequence of achalasia with reported risks ranging from 0.4%-9.2%[58]. One meta-analysis found a risk of SCC of 308.1 per 1000000 per year[59]. One study found that 8.4% of 331 patients with achalasia developed Barrett’s esophagus after undergoing pneumatic dilation[60]. While there are no established guidelines for cancer screening in patients with achalasia, some studies have suggested 3-year interval screening for patients with achalasia for 10 or more years[58].
Given the association between achalasia and esophageal cancer, enhanced imaging in high-risk patients should have value and applications of AI in this population are warranted.
POST CAUSTIC INGESTION AND AI
In the United States, there were over 17000 cases of caustic injury which accounted for about 9% of poisoning cases[61]. Endoscopy has been determined to be an important part of diagnosis and prognosis for these cases of post-caustic ingestion[62,63]. Typically, the Zargar classification is used to help guide evaluation with patients graded 0 through IV. Those with grade III or above typically had complications or death[64]. Artificial intelligence in endoscopy and the role for post-caustic ingestion has not been evaluated. It is reasonable to postulate that with advances in other areas of upper endoscopy in evaluation of the GI lumen for precancerous lesions, achalasia, esophageal carcinoma that there is a role for evaluation of the GI lumen for grading of caustic injury. Further studies are necessary to evaluate whether there is a role for AI assistance in evaluation and if there would be a significant difference in patient outcomes after implementation.
AI AND ESOPHAGEAL SQUAMOUS CELL CARCINOMA
Esophageal cancer has been a large area of investigation due the aggressive disease course and high morbidity and mortality outcomes. It has been reported to be as high as the eighth most common cancer and sixth leading cause of cancer-related death world-wide[65]. As of 2020, there are higher risk geographic areas of concern regarding esophageal cancer in South-Central Asia being the third overall leading cause of cancer-related mortality in males and in the region of Eastern and Southern Africa esophageal cancer ranks second and third in male cancer-mortality respectively. Eastern Africa is also the third leading cause of female related cancer incidence and mortality[66].
Of the two major subtypes of esophageal cancer esophageal squamous cell carcinoma (ESCC) is the predominant histological type world-wide[67]. Classically, ESCC has been associated with risk factors including gender, race, tobacco and alcohol consumption, diet and nutrient intake[67]. Recently, poor oral health and microbiome changes have been associated with the development or predisposition of ESCC[68,69]. By the time of diagnosis of ESCC, disease course is typically found at an advanced stage and often requires highly invasive treatment contributing to poor prognosis, morbidity, and mortality rates. Investigation into early screening is critical, but as with implementation of any mass screening, the method must be evaluated for the benefit of screening tests to reduce cancer vs the risk of over-diagnosing and putting patients through high-risk procedures. It should be noted that there may be specific benefits in implementation of screening in high-risk populations and geographic areas in areas of Africa and Asia. Being an area with high rates of esophageal and gastric cancer, a research study across seven cities in the Henan Province of China enrolled 36154 people for screening using endoscopy and biopsy[70]. They found 46% of patients had precancerous lesions, 2.42% had confirmed cancer. Of those with this confirmed cancer diagnosis, 84% of them had an early stage that underwent prompt treatment with a success rate of 81%. Their study concluded that early detection was crucial in reducing their rate of esophageal and gastric carcinoma in that region[70].
Early-stage detection of ESCC
Early detection is important for improving outcomes for ESCC. Historically, conventional white light endoscopy with biopsy was the gold standard for diagnosis of esophageal cancer[71]. The limitation of this for ESCC is that clinical suspicion needs to be high to perform the procedure and the cancer must be of significant size to be identified on endoscopy. The emergence of chromoendoscopy, using chemicals such as iodine, allowed a staining technique to better detect ESCC. But this procedure can often cause irritation in patients due to mucosal irritation to the GI tract and it increases procedural time per patient.
Alternatively, the emergence of narrow band imaging offers an image-enhancing technique using wavelength filters to observe mucosal differences and vascular patterns on the GI tract that correlates with esophageal cancer (among other uses stated throughout this article). The downside of NBI is that detection rate is dependent on endoscopist experience and subject-ability in processing the information given[71]. Despite these methods, a large multi-center retrospective cohort study by Rodríguez de Santiago et al[35] analyzed over 123000 patients undergoing EGD and found a miss rate of esophageal cancer of 6.4% with a follow-up diagnosis made within 36 mo by repeat endoscopy. This miss rate was present regardless of histologic subtype of esophageal adenocarcinoma or ESCC. Their analysis found that less experienced endoscopists and smaller lesions were associated with the missed detection. Their study acknowledges that there was a low use of chromoendoscopy due to small proportion of early neoplasms across the study and a lack of digital chromoendoscopy at their institutions at the time of the study which may limit applicability[35]. But this still suggests conventional techniques have higher miss rates and newer technology or innovative technique development are essential in assisting and creating a better standard for ESCC detection and to provide a basis for better screening in this aggressive disease.
AI systems – early detection, screening, surveillance
The use of endoscopic AI has recently showed potential to change the diagnostic evaluation for many different gastrointestinal tract diseases. Due to the novelty, ESCC guidelines for use of AI in clinical practice is still being determined.
The use of AI specifically in high-risk populations, may provide great utility to reduce rates of ESCC. Early detection through AI has shown promise through early studies. Ohmori et al[72] used a CNN and showed an accuracy of the AI system for diagnosing ESCC was comparable to that of experienced endoscopists. The system achieved a 76% PPV for detection using non-magnified images and in the differentiation of ESCC using magnified images. Horie et al[73], one of the pioneer investigators of AI in GI endoscopy used a CNN-based AI system to detect ESCC. Their study results showed that their CNN took only 27 s to analyze 1118 images and correctly detected esophageal cancer cases with 98% sensitivity[73]. Thus, it is reasonable that beyond the use of AI systems for evaluation for high-risk patients, at a population-based level, AI systems could be utilized to analyze endoscopic images of patients of medium to low risk that are undergoing EGD for other reasons.
A study by Cai et al[74] specifically developed and validated a computer-aided detection using a DNN to be used for screening for early ESCC. Out of 1332 abnormal and 1096 normal images from 746 patients, they compared their system to 16 endoscopists of various experience levels. Their results showed that the DNN-CAD had an accuracy of 91% compared to their senior endoscopist of 88% and junior endoscopists of 77%. More importantly, after taking the results separately, they allowed the endoscopists to refer to the data and this improved the average diagnostic ability of the endoscopists from an overall average accuracy from 81 to 91%, sensitivity from 74 to 89%, and NPV from 79 to 90%[74].
Depth of invasion
Beyond identifying ESCC at a superficial level for diagnosis, the ability to accurately assess the depth of invasion is important, because it best guides intradisciplinary treatment options[75]. Criteria for diagnosis can be divided into two broad categories: non-magnified endoscopy and magnified endoscopy[75]. In non-magnified endoscopy, macroscopic identifiers are observed such as protrusions and depressions. Magnified endoscopy observes the blood vessel patterns using narrow-based imaging or blue laser imaging; criteria of invasion up to 200 μm (SM1) are candidates for resection because of their lower risk of metastasis[75]. Alternatively, SM2-3 are considered higher risk of metastasis and require consideration for esophagectomy[75]. This diagnostic identification is shown to have endoscopist variability.
The AI systems using CNN have recently emerged to assist the endoscopist and create a higher standard for depth of invasion detection to match or have higher rates than those of expert endoscopists. Evidence was shown by Tokai et al[76], where they used a CNN to differentiate between SM1 and SM2. This was a retrospective study, and 1791 test images were prepared and reviewed by the CNN compared with review by 13 expert endoscopists and found that the AI system demonstrated higher diagnostic accuracy for invasion depth than those of endoscopists.
To determine clinical application from still-images to video, a more recent study by Shimamoto et al[77] utilized real-time assessment of video images for ESCC and compared their AI model with those of expert endoscopists and found that accuracy, sensitivity, and specificity with non-magnified endoscopy were 87%, 50%, and 99% for the AI system and 85%, 45%, 97% for the experts. Accuracy, sensitivity, and specificity with magnified endoscopy was 89%, 71%, and 95% for the AI system and 84%, 42%, 97% for the experts. This suggests that with more inexperienced endoscopists, AI can offer a similar or even higher standard and allow for better patient outcomes with higher depth of invasion diagnosis.
Newer advances in the field of endoscopic AI may offer the potential for diagnosis without biopsy. The Japan esophageal society introduced a classification system for endoscopic diagnosis of ESCC by analyzing intrapapillary capillary loops which help estimate depth of invasion and make a visual diagnosis for ESCC. Although this classification can be endoscopist-dependent, in combination with AI systems, study by Zhao et al[78] used a computer assisted model to allow objective image evaluation and assist in classification of EPCLs and found that their model was 89% accurate in diagnosing the lesion. This was in comparison to accuracy of 92% by senior endoscopists (greater than 15 years), 82% by mid-level endoscopists (10-15 years), and 73% by junior endoscopists (5-10 years). While it is likely not to replace histopathological confirmation, being able to diagnose at a high rate could help more efficiently allocate resources and provide faster diagnosis to help guide clinical intervention in this highly aggressive disease.
In summary, implementation of any cancer-screening for primary prevention is going to require careful analysis of risk-benefits through large-scale medical studies. It is clear that ESCC has a significant presence world-wide and of particular healthcare burden in geographic areas of Africa and Asia. ESCC studies have suggested that implementation of screening can benefit high-risk populations in these areas. AI in endoscopy has emerged with promise in showing consistent results in both early detection, quicker diagnosis, and non-inferior rates of success for the studied patients. Implementation of AI with endoscopic screening of high-risk populations for ESCC should be considered in the coming years as the technology becomes more widely available.
FUTURE PERSPECTIVES FOR AI AND ESOPHAGEAL DISEASES AND MICROBIOME
Eosinophilic esophagitis (EoE)
Eosinophilic esophagitis is a food allergen-mediated inflammatory disease affecting the esophagus. It is traditionally associated with atopic conditions such as asthma and atopic dermatitis[79]. Treatment includes food-elimination diets, proton-pump inhibitors, and topical steroids[79].
Initial diagnosis of eosinophilic esophagitis (EoE) involves mucosal biopsy demonstrating > 15 eosinophils per high-powered field (400× magnification)[79]. In addition to this peripheral eosinophil count (PEC), other histological features may be present in EoE, and can be used to characterize the disease state and to assess for response to therapy, including epithelial thickness, eosinophilic abscess, surface layering, and epithelial alteration[80]. These features have been used to develop a histologic scoring system for diagnosis, the EoEHSS[80]. Both PEC and EoEHSS are evaluated by a pathologist, and are time-consuming processes. EoEHSS additionally requires training and there appears to be inter-observer variability. The need for a more precise and automated process has let to machine learning approaches. Several groups have developed platforms for automated analysis of biopsy images that utilized a deep-convolutional neural network approach to distinguish downscaled biopsy images for features of EoE[81,82]. One platform was able to distinguish between normal tissue, candidiasis, and EoE with 87% sensitivity and 94% specificity. Another platform was able to achieve 82.5% sensitivity and 87% specificity in distinguishing between EoE and controls, despite the potential limitations of image downscaling[82].
In addition to improving efficiency and precision of current diagnostic methods for EoE, AI is a promising tool for the development of new diagnostic methods to subclassify disease and guide treatment. One approach is through evaluation of tissue mRNA expression for unique factors that can classify or subclassify EoE. One group used mRNA transcript patterns to develop a probability score for EoE, in comparison to GERD and controls[83]. This diagnostic model was found to have a 91% diagnostic sensitivity and 93% specificity[83]. Additionally, this EoE predictive score was able to demonstrate response to steroid treatment[83]. Further work may develop new diagnostic criteria, methods for subclassification of disease, and to assess for various therapeutic options.
Esophageal microbiome
Current understanding of the commensal microbiome has developed through various techniques, including 16s rRNA sequencing to describe genus-level composition or shotgun sequencing to describe strain-level composition of a sample microbial community[84]. Various ML models, specifically DL, have been utilized to develop descriptive techniques, disease prediction models based on composition and for exploration of novel therapeutic targets[85].
Initial work on the esophageal microbiome described two compositional types: Type I, associated with the healthy population, mainly consisting of gram-positive flora, including Streptococcus spp., and a Type II, associated with GERD and BE, with higher prevalence of gram-negative anaerobes[86]. Later work stratified esophageal microbiome communities into three types, a Streptococcus spp. predominant (Cluster 2), Prevotella spp. predominant (Cluster 3), and an intermediate abundance type (Cluster 1)[87]. Further work has identified specific flora or groups of flora associated with various disease states as well as a gradient of composition from proximal to distal esophagus[69].
The ML models can be used to expand on this work using both supervised and unsupervised methods. Random Forest classifiers and Least Absolute Shrinkage and Selection Operator feature selection have been used to analyze shotgun genomics data and classify disease state and stage several GI disorders, including colorectal cancer and Crohn’s disease[87-90]. In addition to descriptive methods, machine learning has been used to develop models to predict disease progression in primary sclerosing cholangitis[91]. Finally, correlation-based network analysis methods have been used to assess response to intervention, such as symptomatic response to probiotics and association with microbial changes[92]. Within esophageal disease, a neural network framework has been used to develop a microbiome profile for classification of phenotypes, including datasets from patients with BE and EAC[93]. Future work has the potential to further develop microbiome-based models for detection, assessment of progression, and development of new therapeutics for several esophageal disease states.
DISCUSSION
The emerging use of AI in medicine has the potential for practice changing effects. During the diagnostic process, better visualization techniques, including CAD can assist endoscopists in detection of lesions[94]. When malignancy is detected, AI can be used to predict extent of disease[94]. Following diagnosis, CNN can be used to predict response to treatment as well as risk of recurrence[94].
Of the multiple AI techniques with demonstrated use, some are more likely to be more adaptable to everyday use by clinicians. AI-assisted endoscopy is already being utilized in the area of colorectal disease, with products available on the market to assist with adenoma detection rate and early detection[95]. Given the compatibility of AI solutions with current endoscopic devices, it is likely that broader applications of these systems to other areas of the GI tract are approaching[96].
Some limitations exist in the use of AI-based techniques. First, the quality and number of learning samples significantly affects the accuracy of predictive algorithms. This primarily affects supervised learning networks, where the use of labeled sample data affects the quality of training, and can affect overall accuracy. This concept is sometimes referred to as "garbage in, garbage out." For example, in the detection of gastric cancer, supervised learning algorithms that rely heavily on the quality and quantity of samples may interpret poor images of intestinal metaplasia or atrophy as GC and are heavily data dependent[24]. Semi-supervised and unsupervised learning networks are potential alternatives as they are not entirely data dependent[19]. Another possible limitation is the role of confounding factors- lack of population diversity in training models may lead to lack of generalizability of AI systems to alternate populations.
Finally, privacy will be important to maintain when translated to clinical practice, in both the improvement of training models as well as in patient care. Further legislative discussion is needed to ensure adequate privacy when patient medical data is used and potentially shared for use in ongoing training of AI models[97]. Additionally, this further digitization and storage of patient data will require appropriate security within adapting healthcare system infrastructures[97,98].
CONCLUSION
Clearly, the rapidly developing application of artificial intelligence has shown its wide applicability in gastroenterology and continues to be investigated for the accuracy in endoscopic diagnosis of esophageal and gastric diseases. The esophagogastric diseases including gastric polyps, gastric cancer, BE, achalasia, post-caustic ingestion, ESCC, eosinophilic esophagitis have distinct features that AI can be utilized. The current systems propose a sound base for an AI system that envelops all the esophagogastric diseases. Although this area of active research is very encouraging, further work is needed to better define the specific needs in assessing disease states as well as the cost effectiveness before incorporating AI as a standard tool for daily practice.
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: American College of Gastroenterology; American Society for Gastrointestinal Endoscopy.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): E
P-Reviewer: Fakhradiyev I, Kazakhstan; Liu Y, China S-Editor: Liu JH L-Editor: A P-Editor: Liu JH