Published online Apr 28, 2022. doi: 10.35711/aimi.v3.i2.42
Peer-review started: December 19, 2021
First decision: February 10, 2022
Revised: February 22, 2022
Accepted: April 27, 2022
Article in press: April 27, 2022
Published online: April 28, 2022
Processing time: 129 Days and 22.6 Hours
The pandemic outbreak of the novel coronavirus disease (COVID-19) has highlighted the need to combine rapid, non-invasive and widely accessible techniques with the least risk of patient’s cross-infection to achieve a successful early detection and surveillance of the disease. In this regard, the lung ultrasound (LUS) technique has been proved invaluable in both the differential diagnosis and the follow-up of COVID-19 patients, and its potential may be destined to evolve. Recently, indeed, LUS has been empowered through the development of automated image processing techniques.
To provide a systematic review of the application of artificial intelligence (AI) technology in medical LUS analysis of COVID-19 patients using the preferred reporting items of systematic reviews and meta-analysis (PRISMA) guidelines.
A literature search was performed for relevant studies published from March 2020 - outbreak of the pandemic - to 30 September 2021. Seventeen articles were included in the result synthesis of this paper.
As part of the review, we presented the main characteristics related to AI techniques, in particular deep learning (DL), adopted in the selected articles. A survey was carried out on the type of architectures used, availability of the source code, network weights and open access datasets, use of data augmentation, use of the transfer learning strategy, type of input data and training/test datasets, and explainability.
Finally, this review highlighted the existing challenges, including the lack of large datasets of reliable COVID-19-based LUS images to test the effectiveness of DL methods and the ethical/regulatory issues associated with the adoption of automated systems in real clinical scenarios.
Core Tip: Challenging coronavirus disease 2019 (COVID-19) pandemic through the identification of effective diagnostic and prognostic tools is of outstanding importance to tackle the healthcare system burdening and improve clinical outcomes. Application of deep learning (DL) in medical lung ultrasound may offer the advantage of combining non-invasiveness and wide accessibility of ultrasound imaging techniques with higher diagnostic performance and classification accuracy. This paper overviews the current applications of DL models in medical lung ultrasound imaging in COVID-19 patients, and highlight the existing challenges associated with the effective clinical application of automated systems in the medical imaging field.
- Citation: De Rosa L, L'Abbate S, Kusmic C, Faita F. Applications of artificial intelligence in lung ultrasound: Review of deep learning methods for COVID-19 fighting. Artif Intell Med Imaging 2022; 3(2): 42-54
- URL: https://www.wjgnet.com/2644-3260/full/v3/i2/42.htm
- DOI: https://dx.doi.org/10.35711/aimi.v3.i2.42
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a life-threatening infectious virus and its related disease (COVID-19) represents a still ongoing challenge for humans. At time of writing, over 497 million infections have been recorded worldwide including more than 6.1 million attributable deaths[1]. Despite the large number of vaccination programs introduced from the end of 2020 has represented an opportunity to minimise the risk of severe COVID-19 and death, the spread of new genetic viral variants with a higher probability of contagion has raised a renewed strong concern for either not vaccinated and vaccinated people. Thus, since the outbreak of the pandemic, research has continuously looked for a quick and reliable way to diagnose the disease, treat and monitor people affected by coronavirus.
To date, molecular test based on real time quantitative reverse transcription polymerase chain reaction (RT-qPCR) assay by nasopharyngeal swabs along with the serological antibody-detecting and antigen-detecting tests are the current accepted diagnostic tools for the conclusive diagnosis of COVID-19[2]. RT-qPCR may take up to 24 h to provide information and requires multiple tests for definitive results and, in addition, it is not relevant to assess the disease severity. Furthermore, the accuracy of molecular and serological tests remains highly dependent on timing of sample collection relative to infection, improper sampling of respiratory specimens, inadequate preservation of samples and technical errors, particularly contamination during RT-qPCR process and cross-reactivity in the immunoassay[3,4].
To complement conventional in vitro analytical techniques of COVID-19, biomedical imaging techniques have demonstrated great potential in clinical diagnostic evaluation by providing rapid patient assessment in the presence of high pre-test probability. Furthermore, imaging techniques are currently important in the follow-up of subjects with COVID-19[5,6]. Among the imaging techniques, chest computed tomography (CT) is considered the primary diagnostic modality and an important indicator for assessing severity and progression of COVID-19 pneumonia[7,8], although it has been reported to have limited specificity[9-11]. Indeed, the CT imaging features can overlap between COVID-19 and other viral pneumonia. Moreover, CT scanning is expensive, not easy to perform in the COVID-19 context, and multiple risks are associated with it, such as radiation exposure and cross-infection risk associated with repeated use of a CT suite[12], along with unavailability of CT in many parts of the world.
In the last few years, lung ultrasound (LUS) technique has become increasingly popular and a good option for real-time point-of-care testing, with several advantages making it a valuable tool in the fight against COVID-19[13], although it has specificity limits comparable to those of chest CT.
Ultrasound (US) is a low-cost, non-radioactive medical imaging method, particularly indicated for evaluation in pregnant women and children, which is portable to the bedside or patient’s home and is easy to sterilise. Moreover, the risk of COVID-19 cross-infection can be limited by making use of disposable ultrasound gel with a portable probe[14]. In addition, some studies indicate that LUS shows excellent performances in speed of execution and accuracy of diagnosis in case of respiratory failure[15]. Furthermore, compared with chest X-ray, LUS demonstrated higher sensitivity in detecting pneumonia[16] and similar specificity in the diagnosis of pneumothorax[15]. On the other hand, the distinctive LUS features (B-lines, consolidations, pleural thickening and rupture) observed in patients with varying severity of COVID pneumonia are similar to the features seen in patients with pneumonia of different aetiologies. Indeed, a recent review[17] on ultrasound findings of LUS in COVID-19 demonstrated that LUS has high sensitivity and reliability in ruling out lung involvement, but at the expense of low specificity. Therefore, especially in the case of low prevalence of the disease, at present LUS cannot be considered a valid gold standard in clinical practice.
Ultrasound image processing techniques have assumed great importance in recent years, with the growing experience that accurate image processing can significantly help in extracting quantitative characteristics to assess and classify the severity of diseases. Accordingly, sophisticated techniques of automated image processing, that include the use of artificial intelligence (AI) methods, have been developed and applied to assist LUS imaging in the detection of COVID-19 and make such assessment more objective and accurate. AI methods - from machine learning (ML) to deep learning (DL), indeed, aim to imitate cognitive functions and stand out in automatically recognizing complex patterns in imaging data, providing quantitative rather than qualitative assessments. The primary purpose of applying AI methods in medical imaging is to improve the visual recognition of certain features in images to produce lower-than-human error rates. Furthermore, an enhancement in LUS performance can reduce the use of more invasive and time-consuming techniques, facilitating both faster diagnosis and recognition of earlier stages of the disease[18]. To allow a quick development of highly performant AI models, a large amount of accessible and validated data to train and test AI models is a critical requirement that can be achieved, for instance, with the development of shared big data archives. Indeed, one of the most common problems associated with using limited training samples is the over-fitting of DL models. To address this issue, two main approaches can be selected: model optimization and transfer learning. These strategies significantly improve the performance of DL models. Likewise, data pre-processing and data augmentation/enhancement can be useful additional strategies[19,20].
The most common applications of DL methods in clinical imaging, and hence in medical ultrasound imaging as well, are object detection, object segmentation, and object classification[21]. The main architectures applied in current analysis are convolutional neural networks (CNNs) and recurrent neural networks (RNNs)[22]. CNNs are architectures able to work with 2D and 3D input images and RNNs recognize the image's sequential characteristics and use patterns to predict the next likely scenario[23].
Since the outbreak of the pandemic, many proposals have been made based on AI methods applied to LUS scans of COVID-19 patients. Here we propose a comprehensive systematic review of the literature on the use of AI technology, DL in particular, to aid in the fight against COVID-19.
A literature search to identify all relevant articles on the use of DL tools applied to LUS imaging in patients affected by COVID-19 virus was conducted.
This systematic review was carried out using the PubMed/Medline electronic database and according to the preferred reporting for systematic reviews and meta-analysis (PRISMA) guidelines[24,25]. We performed a systematic search covering the period from March 2020 (from the outbreak of the pandemic) to 30 September 2021. The search strategy was restricted to English-language publications.
We performed an advanced research concatenating terms with Boolean operators. In particular, search words and key terms used in the search included ("lung ultrasound" OR "lus") AND ("COVID-19" OR "coronavirus" OR "SARS-CoV2") AND ("artificial intelligence" OR "deep learning" OR "neural networks" OR "CNN").
The inclusion criteria were: Studies that include COVID-19 patients with LUS acquisitions and developed or tested DL-based algorithms on LUS images or on features extracted from the images; No restriction on the ground truth adopted to analyse the presence/absence of COVID-19 and/or the severity of lung disease (e.g., PCR, visual evaluation of video/images and score assignment by expert clinicians); No restriction on the type of DL architecture used in the studies. Studies on paediatric population were excluded. Studies were restricted to peer reviewed articles and conference proceedings. However, the following publication types were excluded: reviews and conference abstracts.
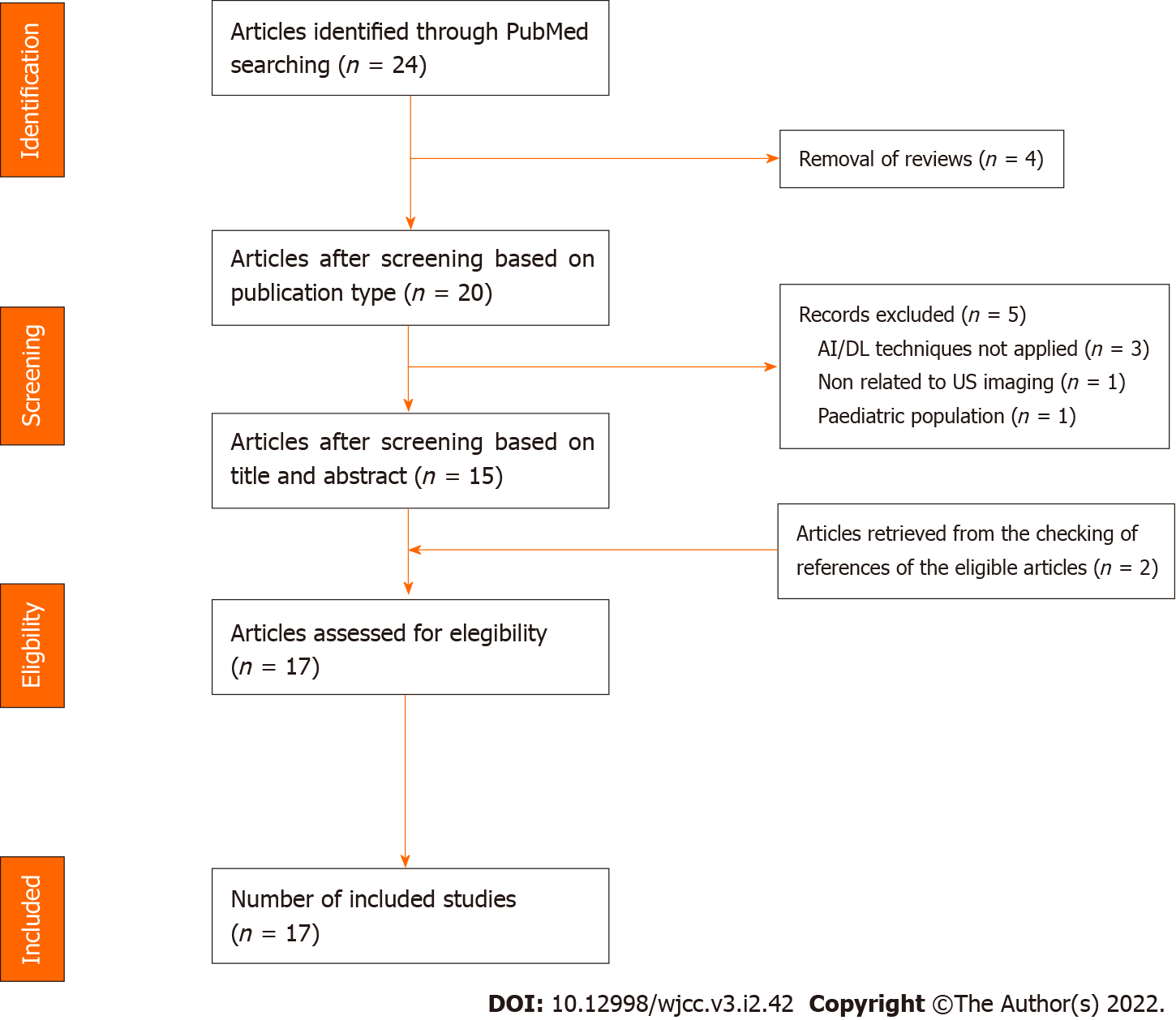
Two investigators (DRL and FF) screened the articles independently. Disagreement between reviewers was resolved by consensus via discussion. The reasons for the exclusion of some trials are described in the Results section. Publications by the same research group or by different groups using the same dataset were included in the analysis. After the selection of the articles, we collected the following characteristics: First author’s surname, date of publication, sample size, general characteristics of the study populations, AI techniques used, validation methods and main results obtained. The study selection process is presented in Figure 1.
Twenty-four articles resulted after querying the database and screened for eligibility (Figure 1). Of the 24 articles, we discarded four references as review papers. After examining the titles and abstracts, we excluded five articles: one manuscript did not include DL methods applied on US imaging, three papers were not based on AI and DL approaches, and one article was focused on the paediatric population. Moreover, two additional papers, retrieved from the checking of references of the eligible articles, were included. Finally, 17 articles[26-42] were selected for full-text screening and included in our analysis (Table 1 and 2). The following part of the section provides a concise overview of the studies’ main features.
| Ref. | Publication date | Journal | Sample size1, N° pts/videos/images | Subjects | Main results |
| Arntfield et al[26] | 22/02/2021 | BMJ Open | 243/612/121k | COVID +, COVID -, HPE | Overall Acc = 0.978AUC = 1/0.934/1 for COVID +, COVID -, HPE |
| Awatshi et al[27] | 23/03/2021 | IEEE Trans Ultrason Ferroelectr Freq Control | -/64/1.1k | COVID +, Healthy, PN | 5-fold validation: Acc = 0.829 |
| Barros et al[28] | 14/08/2021 | Sensors | 131/185/- | COVID +, PN bacterial, Healthy | Best model (Xception+LSTM): Acc = 0.93 – Se = 0.97 |
| Born et al[29] | 12/01/2021 | Applied Sciences | 216/202/3.2k | COVID +, Healthy, PN | External validation: Se = 0.806 – Sp = 0.962 |
| Born et al[30] | 24/01/2021 | ISMB TransMed | -/64/1.1k | COVID +, Healthy, PN | Overall Acc = 0.89Binarization COVID y/n: Se = 0.96 – Sp = 0.79 – F1score = 0.92 |
| Chen et al[31] | 29/06/2021 | IEEE Trans Ultrason Ferroelectr Freq Control | 31/45/1.6k | COVID-19 PN | 5-fold validation: Acc = 0.87 |
| Dastider et al[32] | 20/02/2021 | Comput Biol Med | 29/60/14.3k | COVID-19 PN | Independent data validation: Acc = 0.677 – Se = 0.677 – Sp = 0.768 – F1score = 0.666 |
| Diaz Escobar et al[33] | 13/08/2021 | PLos One | 216/185/3.3k | COVID +, PN bacterial, Healthy | Best model (InceptionV3): Acc = 0.891 – AUC = 0.971 |
| Erfanian Ebadi et al[34] | 04/08/2021 | Inform Med Unlocked | 300/1.5k/288k | COVID +, PN | 5-fold validation: Acc = 0.90 – PP=0.95 |
| Hu et al[35] | 20/03/2021 | BioMed Eng OnLine | 108/-/5.7k | COVID + | COVID detection: Acc = 0.944 – PP = 0.823 – Se = 0.763 – Sp=0.964 |
| La Salvia et al[36] | 03/08/2021 | Comput Biol Med | 450/5.4k/> 60k | Hospitalised COVID-19 | External validation (ResNet50): Acc = 0.979 – PP=0.978 – F1score = 0.977 – AUC = 0.998 |
| Mento et al[37] | 27/05/2021 | J Acoust Soc Am | 82/1.5k/315k | COVID-19 confirmed | % Agreement DL and LUS = 96% |
| Roy et al[38] | 14/05/2020 | IEEE Trans | 35/277/58.9k | COVID-19 confirmed, COVID-19 suspected, Healthy | Segmentation: Acc = 0.96 – DICE = 0.75 |
| Sadik et al[39] | 09/07/2021 | Health Inf Sci Syst | -/123/41.5k | COVID +, PN, Healthy | COVID y/n (VGG19+SpecMen): PP = 0.81 – F1score = 0.89 |
| Muhammad et al[40] | 25/02/2021 | Information Fusion | 121 videos + 40 frames | COVID +, PN bacterial, Healthy | Overall: Acc = 0.918 – PP = 0.925 |
| Tsai et al[41] | 08/03/2021 | Phys Med | 70/623/99.2k | Healthy, Pleural effusion pts | Pleural effusion detection:Acc = 0.924 |
| Xue et al[42] | 20/01/2021 | Med Image Anal | 313/-/6.9k | COVID-19 confirmed | 4-level and binary disease severity:Acc = 0.75 and Acc = 0.85 |
| Ref. | DL architecture | Input of DL models | Available dataset | Available code | Pre-trained/TL | Test independent | Data Augmentation | Explainability |
| Arntfield et al[26] | CNN | SF | No | Yes (on github) | Yes | Yes | Yes | Yes |
| Awatshi et al[27] | CNN | SF | No | Yes (on github) | Yes | No (five-fold) | Yes | Yes |
| Barros et al[28] | CNN+LSTM | SF | Yes | Yes (on github) | Yes | No(five-fold) | No | Yes |
| Born et al[29] | 3D CNN | MF | Yes | Yes (on github) | Yes | No(five-fold) | Yes | Yes |
| Born et al[30] | CNN | SF | Yes | Yes (on github) | Yes | No(five-fold) | Yes | No |
| Chen et al[31] | MLFCNN | SF | No | Yes (on github) | No | No(five-fold) | No | No |
| Dastider et al[32] | CNN+LSTM | SF | No | Yes (on github) | Yes | No(five-fold) | Yes | Yes |
| Diaz Escobar et al[33] | CNN | SF | No | No | Yes | No(five-fold) | Yes | No |
| Erfanian Ebadi et al[34] | 3D CNN | MF | No | Yes (on github) | Yes | No(five-fold) | No | Yes |
| Hu et al[35] | CNN + MCRF | SF | No | No | Yes | Yes | Yes | Yes |
| La Salvia et al[36] | CNN | SF | No | No | Yes | Yes | Yes | Yes |
| Mento et al[37] | CNN+ STN | SF | No | No | No | - | No | No |
| Roy et al[38] | CNN+ STN | SF | Yes (on request) | Yes (on github) | No | Yes | Yes | Yes |
| Sadik et al[39] | CNN | SF | No | No | Yes | Yes | Yes | Yes |
| Muhammad et al[40] | CNN | SF | Yes | No | No | No(five-fold) | Yes | Yes |
| Tsai et al[41] | CNN+ STN | MF | No | No | Yes | No(ten-fold) | No | No |
| Xue et al[42] | CNN | SF | No | No | No | Yes | Yes | Yes |
Authors of seven[27-30,33,39,40] of the seventeen selected articles (41.2%) extrapolated their datasets from the free access LUS database acquired by point-of-care ultrasound imaging and made available firstly by Born et al[30]. Instead, an Italian group firstly introduced the Italian COVID-19 Lung Ultrasound DataBase (ICLUS-DB)[38], which is accessible upon mandatory request to the authors, and that was used in two other studies[32,37]. Noteworthy, Roy et al[38] have created a platform through which physicians can access algorithms, upload their data and see the algorithm's evaluation of the data.
Besides dataset open access, access to the code for the neural network is also important to reproduce results and compare performances. Seven articles[26-30,32,38] (41.2%) made the source code implementing the proposed DL architecture available for download from the Git-hub repository.
In the majority of the selected papers, DL architectures work with single frame images as input and only three publications[29,34,41] (17.6%) report DL architectures based on image sequences (i.e., video). However, six studies[28,30,32,37-39] (35.3%), despite adopting a DL architecture designed to perform single-frame classification, also propose additional methods to fulfil video-based classification. In particular, Roy et al[38] proposed an aggregation layer system of frame-level scores to produce predictions on LUS videos and Mento et al[37] proposed an alternative video-based classification using a threshold-based system on the frame-level scores obtained from DL architecture.
Other authors[32] adopted a Long Short-Term Memory (LSTM) system, which has been used to exploit temporal relationships between multiple frames by taking long time series as input, over performing their results obtained by CNN without LSTM.
Finally, Xue et al[42] applied AI models for patient-level assessment of severity using a final module across the entire architecture that works with ML rather than DL systems.
The proposed DL models have been tested on a database entirely independent from the training database in seven articles[26,35-39,42] (41.2%); five-fold and ten-fold cross-validation techniques were applied in nine[27-34,40] (52.9%) and one[41] (5.9%) studies, respectively. Among the papers that tested DL models on an independent database, the percentage of data used for the testing ranged from 33%[35] to 20%[38] and 10%[26,36] of the overall data. Born et al[29], alongside the five-fold cross-validation technique in the training/test phase of the DL model, also used an independent validation dataset made-up of 31 videos (28 convex and 3 linear probes) from six patients. Indeed, Roy et al[38], for instance, used 80 videos/10709 frames out of the total 277 videos/58924 frames to test their DL model.
In all studies, the splitting of data between training set and test set was performed either at the patient-level or at the video-level. Thus, all the frames of a single video clip belonged either to the training or to the test set.
Twelve (70.6%) research groups extended their LUS database by augmentation. The main strategies for data augmentation applied to LUS images were: Horizontal/vertical flipping[26,27,29,30,32,33,36,38-40,42], bidirectional arbitrary rotation[26,27,29,30,32,33,35,38-40,42], horizontal and vertical shift[30,32,38,39,42]; filtering, colour transformation, adding salt and pepper noise, Gaussian noise[36,38,42], normalisation of grey levels’ intensity[38]. Although proposed by all the authors, only seven papers[26,29,30,32,33,38,40] provided details on the amplitude of image rotation. In particular, Dastider et al[32] applied rotations in the range of 0 ± 360 degrees, while other authors have limited image rotations to 10 degrees[26,29,30,33], ± 15 degrees[38] and ± 20 degrees[40], respectively. The remaining five papers[28,31,34,37,41] (29.4%) did not perform data augmentation.
Among the selected articles, tools for interpreting the network output were provided in twelve studies (70.6%), whereas in the remaining five (29.4%) the DL algorithms’ outcomes were proposed as black box systems. The majority of papers[26-29,32,35,36,38,40] reported the Gradient-weighted Class Activation Mapping (Grad-CAM) as the preferred explainability tool. Grad-CAM uses gradients to create a location map to highlight the region of interest of the images[43]. Instead, Sadik et al[39] used a colormap jet to visualise a heat map overlay to US images; Erfanian Ebadi et al[34] adopted an activation map system to detect and segment features in LUS scans. Furthermore, one study[42] showed LUS images with overlaid colormaps to indicate the segmentation zone of ultrasound according to the different severity. Roy et al[38], differently, provided an ultrasound colormap overlay on the LUS frame/video and used four colours to distinguish the different classes of disease severity recognized by DL architecture.
Most of the selected papers applied the AI system to diagnose COVID-19 and/or discriminate between COVID-19 and other lung diseases (such as bacterial pneumonia)[26-30,33,34,39,40]. The first approach using DL architecture for automatic differential diagnosis of COVID-19 from LUS data was POCOVID-Net[30].
However, a fair number of studies have focused on assessing the severity of COVID-19[31,32,35-38,42]. In particular, a disease severity score is assigned to the single image according to some characteristics visible in the image pattern. Most of the articles used four severity classes by assigning a score to the single frame from 0 to 3[31,32,35-38], as defined by Soldati et al[44]. Xue et al[42] proposed a classification in five classes of pneumonia severity (score from 0 to 4) along with a binary severe/non-severe classification. Furthermore, these authors used the DL technology exclusively to implement a segmentation phase based on a VGG network, while the classification phase still employed a more traditional, features-based machine learning approach. Finally, La Salvia et al[36] proposed a classification based on three severity classes and a modified version considering a seven-classes scenario.
Furthermore, Arntfield et al[26] showed that their network was able to recognize pathological pattern in LUS images with higher sensitivity than sonographers; whilst an InceptionV3 network proposed by Diaz-Escobar et al[33] was able to discriminate COVID-19 pneumonia from healthy lung and other bacterial pneumonia with an accuracy of 89.1% and an area under the ROC curve of 97.1%.
Curiously, one of the eligible papers[41] did not include confirmed cases of COVID-19 patients. The authors’ aim was to design an algorithm capable of identifying the presence of pleural effusion. However, we have included this work in our systematic review, because small pleural effusions are rarely reported in COVID-19 patients. Therefore, the detection of pneumonia with pleural effusion can help rule out the hypothesis of COVID-19 disease.
From our analysis, it emerged that most of the studies have proposed convolutional neural networks (CNNs) as DL models to generate screening systems for COVID-19. In particular, all publications with the exception of one[31] used the CNN network. Conversely, Chen et al[31] developed a multi-layer fully connected neural network for scoring LUS images in assessing the severity of COVID-19 pneumonia.
Among the DL systems included in this review, most of them were generated starting from DL architectures already proposed for other tasks[26-30,32-36,39,42], suitably modified and trained for new tasks. Furthermore, many works compared the results of their architectures with those obtained using existing and well-known architectures[27-30,32,33,35,38-40]. In particular, the following DL architectures were adapted to fulfil the requirements of LUS analysis to assist in COVID-19 detection and/or assessment of the severity of the lung disease, or just to compare their performances: VGG-19[28,33,39] and VGG-50[28-30,33]; Xception[26,28,39]; ResNet 50[27,33,36,40]; NasNetMobile[27,29,39]; DenseNet[32,39].
More in detail, Awasthi et al[27] proposed Mini-COVIDNet, a modified MobileNet model belonging to the CNN’s networks family and originally developed for detecting objects in mobile applications[45]. Barros et al[28], along with their proposed DL model, also investigated the impact of using different pre-trained CNN architectures in extracting spatial features that were successively classified by a LSTM model. Finally, Born et al[29] derived their DL video-based models from a model that was pre-trained on lung CT scans[46].
All aforementioned architectures are pre-trained on ImageNet[47].
Partly due to the recent outbreak of the pandemic and to the difficulty of having standardised high quality archives of US images, only few of the selected studies relied on a large dataset in terms of enrolled patients. Six papers (35.3%) reported a sample size greater than 200 subjects (namely, 243, 216, 216, 300, 450 and 313 in references[26,29,33,34,36,42] respectively).
However, despite the relatively low number of subjects, the total number of LUS videos reaches up to 5400 in one study[36], with an average equal to 1589 videos[26,29,33,34,36]. Among the studies carried out on a low sample size, Dastider et al[32] included 29 patients and 60 videos, whilst 35 patients/45 videos and 35 patients/277 videos were analysed in references Chen et al[31] and Roy et al[38], respectively. However, it should be noted that Roy et al[38] published their work at the beginning of the COVID-19 pandemic, when the total number of COVID-19 patients was still relatively limited. In the paper by Xue et al[42], the number of frames/video was not reported.
The paper reviews the different DL techniques able to work with LUS images in assisting the diagnosis and/or prognosis of the COVID-19 disease published since the outbreak of the pandemic. In the selected documents, the use of DL systems aimed to achieve an accuracy comparable to or better than clinical standards to provide a faster diagnosis and/or follow-up in COVID-19 patients.
Most of the papers present pre-trained DL architectures[26-30,32-36,39,42] that were modified and adapted to new data. This approach is also known as transfer learning (TL) technique - i.e., a training strategy for new DL models with reduced datasets. The network is pre-trained on a very large dataset, such as ImageNet, with millions of images intentionally created to facilitate the training of DL models, focusing on image classification and object location/detection tasks[48]. Indeed, deeper models are difficult to train and provide inconsistent performances when trained on a limited amount of data[49]. Therefore, most of the studies based on DL systems to classify COVID-19 images appropriately use the TL strategy as large datasets of US images from COVID-19 patients are not yet easily available, partly because the coronavirus disease is a relatively recent concern.
Furthermore, most of the proposed systems shared the same design, i.e., CNN’s architectures. CNNs have several applications in medical imaging – among others, image segmentation and object detection[50]. However, CNNs are particularly suited for image classification problems[51] and, consequently, represent an optimal solution for the classification of the disease severity from US images.
To date, one of the main challenges faced by DL architectures applied to LUS images of COVID-19 patients are the limited datasets in the available databases. This problem could benefit from creating open access databases that collect large amounts of data from multiple centres. In some of the selected studies, a first attempt to overcome this issue is evident, with particular emphasis on the work by Born et al[30], the authors who first collected a free access dataset of lung images from healthy controls and patients affected by COVID-19 or other pneumonia.
The development of public and multicentre platforms would guarantee the collection of a continuously growing amount of data, large and highly heterogeneous, suited for the training and testing of new DL applications in medical imaging, both in the COVID-19 and LUS field. Furthermore, this would allow an easier comparison of performances among DL models proposed in different studies. However, alternative approaches are often used in the testing phase that do not require the use of independent data sets to evaluate the performance of the model in the event of a limited number of images available. Among these, the k-fold cross-validation is a statistical method used to evaluate the ability of ML models to generalise to previously unseen data. Despite being widely used in ML models, the k-fold cross validation approach is less reliable than tests performed using an external dataset; the latter is always preferable to test model's ability to adapt properly to new, previously unseen data.
Data augmentation techniques are an alternative strategy to overcome the issue of the limited amounts of data, largely adopted in practice. These techniques generate different versions of a real dataset artificially to both increase its size and the power of model's generalisation. Despite the great advantage in increasing data to feed DL architectures, data augmentation techniques should be used with awareness, as some geometric transformations could be unrealistic when applied to LUS images (e.g., angles of rotations greater than 30°). In the field of DL applied to medical imaging, the use of architectures designed to work with 3D images is another interesting challenge. Indeed, a DL system that operates with 3D data input usually requires a larger amount of data for training, as a 3D network contains a parameters’ number that is orders of magnitude greater than a 2D network. This could significantly increase the risk of overfitting, especially in the case of limited dataset availability. In addition, the training on large amounts of data requires high computational costs associated with memory and performance requirements of the tools used. LUS images are usually recorded in the form of videoclips (2D + time) and can be assimilated to 3D data. Exploitation of dynamic information naturally embedded in image sequences has proven very important in the analysis of lung echoes. In particular, changes induced by COVID-19 viral pneumonia are better detectable in LUS through the analysis of multi-frames acquisition due to its ability in capturing dynamic features, e.g., pleural sliding movements and generation of B-line artefacts[44].
Regardless of the data format (i.e., 3D, 2D or 2D+time images), the labelling of ground truth data is required in supervised DL applications and should be provided by skilled medical professionals. However, it is a time-consuming activity, in particular in the 2D approach that is characterised by a high number of samples.
Indeed, some authors demonstrated that the performance in pleural effusion classification on LUS images obtained with the video-based approach was comparable to that obtained with frame-based analysis, despite a significant reduction in labelling effort[41]. Furthermore, Kinetics-I3D network was able to classify LUS video sequences with great accuracy and efficiency[34]. On the other hand, the video-based approach has also revealed a reduced accuracy in patients classification with respect to the single frame analysis; however, this could be explained by the relatively reduced number of available LUS clips[29].
Extending the use of DL architectures beyond multi-frame analysis with respect to single 2D images is highly desirable. In particular, these methods could be effectively used to assign a patient-level disease severity score. In fact, this information plays a key role in the selection of treatment, monitoring of disease progression and management of medical resources (e.g., mechanical ventilator needed).
Code availability is another very critical issue in applications of AI in medical imaging. Indeed, the lack of ability to reproduce the training of the proposed DL models or to test these models on new US images is a rather widespread problem. Often, authors do not provide access to either the source code used to train NNs or the final weight of the trained network. On the other hand, the availability of this information would greatly facilitate the diffusion of new AI systems in the clinical setting.
DL systems are often presented as black boxes - i.e., they produce a result without providing a clear understanding in "human terms" of how it was obtained. The black-box nature of the algorithms has restricted their clinical use until now. Consistently, the explainability - i.e., making clear and understandable the features that influence the decisions of a DL model - is a critical point to guarantee a safe, ethical, and reliable use of AI. Especially in medical imaging applications, explainability is very important as it gives the opportunity to highlight regions of the image containing the visual features that are critical for the diagnosis. Gradient-weighted Class Activation Mapping (Grad-CAM) is a promising technique for producing "visual explanations" of decisions taken from a large class of CNN-based models, making their internal behaviour more understandable, thus partially overcoming the black-box problem. The basic idea is to produce a rough localization map that highlights the key regions in the image that have a major effect on customization of network parameters, thus maximally contributing to the prediction of outcomes[43].
These maps visualised areas using a blue-to-red scale, with the highest/lowest contribution to the class prediction operated by the model. The clinical use of DL systems is a crucial issue. One of the major current limitations of LUS imaging in COVID patients is the specificity. Focusing the design of DL systems to overcome this limit could really represent a benefit in the clinical setting.
Along this line, some of the included studies tested the agreement between physicians' ability to classify COVID-19 patients and that proposed by neural networks. Furthermore, this finding suggests that the automated system can capture some features (biomarkers) in US images that are not clearly visible to the human eye.
Finally, another important issue to mention is the use of the quantitative evaluation indicators and the analysis of the benchmarking techniques adopted to evaluate the effectiveness of the proposed methods. Unfortunately, the tools examined in the selected manuscripts had very heterogeneous targets (Table 1, Main results column), ranging from diagnostic to prognostic purposes or assessment of disease severity. This dispersion of intent and the few articles published in the literature at present make any comparison or analysis very difficult.
The studies analysed in this article have shown that DL systems applied to LUS images for the diagnosis/prognosis of COVID-19 disease have the potential to provide significant support to the medical community. However, there are a number of challenges to overcome before AI systems can be regularly employed in the clinical setting. On the one hand, the critical issues related to the availability of high-quality databases with large sample size of lung images/videos of COVID-19 patients and free access to datasets must be addressed. On the other hand, existing concerns about the methodological transparency (e.g., explainability and reproducibility) of DL systems and the regulatory/ethical and cultural issues that the clinical use of AI methods raise must be resolved. Finally, a closer collaboration between the communities of informatics/engineers and medical professionals is desirable to facilitate the outcome of adequate guidelines for the use of DL in US pulmonary imaging and, more generally, in medical imaging.
The current coronavirus disease 2019 (COVID-19) pandemic crisis has highlighted the need for biomedical imaging techniques in rapid clinical diagnostic evaluation of patients. Furthermore, imaging techniques are currently important in the follow-up of subjects with COVID-19. The lung ultrasound technique has become increasingly popular and is considered a good option for real-time point-of-care testing, although it has specificity limits comparable to those of chest computed tomography.
The application of artificial intelligence, and of deep learning in particular, in medical pulmonary ultrasound can offer an improvement in diagnostic performance and classification accuracy to a non-invasive and low-cost technique, thus implementing its diagnostic and prognostic importance to COVID-10 pandemic.
This review presents the state of the art of the use of artificial intelligence and deep learning techniques applied to lung ultrasound in COVID-19 patients.
We performed a literature search, according to preferred reporting items of systematic reviews and meta-analysis guidelines, for relevant studies published from March 2020 - to 30 September 2021 on the use of deep learning tools applied to lung ultrasound imaging in COVID-19 patients. Only English-language publications were selected.
We surveyed the type of architectures used, availability of the source code, network weights and open access datasets, use of data augmentation, use of the transfer learning strategy, type of input data and training/test datasets, and explainability.
Application of deep learning systems to lung ultrasound images for the diagnosis/prognosis of COVID-19 disease has the potential to provide significant support to the medical community. However, there are critical issues related to the availability of high-quality databases with large sample size and free access to datasets.
Close collaboration between the communities of computer scientists/engineers and medical professionals could facilitate the outcome of adequate guidelines for the use of deep learning in ultrasound lung imaging.
| 1. | World Health Organization. (2022, April 13). Coronavirus (COVID-19) Dashboard. Available from: https://covid19.who.int. |
| 2. | Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92:1518-1524. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 901] [Cited by in RCA: 1103] [Article Influence: 183.8] [Reference Citation Analysis (0)] |
| 3. | Yang Y, Yang M, Yuan J, Wang F, Wang Z, Li J, Zhang M, Xing L, Wei J, Peng L, Wong G, Zheng H, Wu W, Shen C, Liao M, Feng K, Yang Q, Zhao J, Liu L, Liu Y. Laboratory Diagnosis and Monitoring the Viral Shedding of SARS-CoV-2 Infection. Innovation (N Y). 2020;1:100061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 120] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 4. | Mardian Y, Kosasih H, Karyana M, Neal A, Lau CY. Review of Current COVID-19 Diagnostics and Opportunities for Further Development. Front Med (Lausanne). 2021;8:615099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 5. | Hoffmann T, Bulla P, Jödicke L, Klein C, Bott SM, Keller R, Malek N, Fröhlich E, Göpel S, Blumenstock G, Fusco S. Can follow up lung ultrasound in Coronavirus Disease-19 patients indicate clinical outcome? PLoS One. 2021;16:e0256359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (1)] |
| 6. | Martini K, Larici AR, Revel MP, Ghaye B, Sverzellati N, Parkar AP, Snoeckx A, Screaton N, Biederer J, Prosch H, Silva M, Brady A, Gleeson F, Frauenfelder T; European Society of Thoracic Imaging (ESTI), the European Society of Radiology (ESR). COVID-19 pneumonia imaging follow-up: when and how? Eur Radiol. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, Diao K, Lin B, Zhu X, Li K, Li S, Shan H, Jacobi A, Chung M. Chest CT Findings in Coronavirus Disease-19 (COVID-19): Relationship to Duration of Infection. Radiology. 2020;295:200463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1728] [Cited by in RCA: 1608] [Article Influence: 268.0] [Reference Citation Analysis (3)] |
| 8. | Lyu P, Liu X, Zhang R, Shi L, Gao J. The Performance of Chest CT in Evaluating the Clinical Severity of COVID-19 Pneumonia: Identifying Critical Cases Based on CT Characteristics. Invest Radiol. 2020;55:412-421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 9. | Raptis CA, Hammer MM, Short RG, Shah A, Bhalla S, Bierhals AJ, Filev PD, Hope MD, Jeudy J, Kligerman SJ, Henry TS. Chest CT and Coronavirus Disease (COVID-19): A Critical Review of the Literature to Date. AJR Am J Roentgenol. 2020;215:839-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 122] [Article Influence: 20.3] [Reference Citation Analysis (1)] |
| 10. | Inui S, Gonoi W, Kurokawa R, Nakai Y, Watanabe Y, Sakurai K, Ishida M, Fujikawa A, Abe O. The role of chest imaging in the diagnosis, management, and monitoring of coronavirus disease 2019 (COVID-19). Insights Imaging. 2021;12:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 11. | Xu B, Xing Y, Peng J, Zheng Z, Tang W, Sun Y, Xu C, Peng F. Chest CT for detecting COVID-19: a systematic review and meta-analysis of diagnostic accuracy. Eur Radiol. 2020;30:5720-5727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 131] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 12. | Self WH, Courtney DM, McNaughton CD, Wunderink RG, Kline JA. High discordance of chest x-ray and computed tomography for detection of pulmonary opacities in ED patients: implications for diagnosing pneumonia. Am J Emerg Med. 2013;31:401-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 235] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 13. | Smith MJ, Hayward SA, Innes SM, Miller ASC. Point-of-care lung ultrasound in patients with COVID-19 - a narrative review. Anaesthesia. 2020;75:1096-1104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 239] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 14. | Akl EA, Blažić I, Yaacoub S, Frija G, Chou R, Appiah JA, Fatehi M, Flor N, Hitti E, Jafri H, Jin ZY, Kauczor HU, Kawooya M, Kazerooni EA, Ko JP, Mahfouz R, Muglia V, Nyabanda R, Sanchez M, Shete PB, Ulla M, Zheng C, van Deventer E, Perez MDR. Use of Chest Imaging in the Diagnosis and Management of COVID-19: A WHO Rapid Advice Guide. Radiology. 2021;298:E63-E69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (1)] |
| 15. | Walden A, Smallwood N, Dachsel M, Miller A, Stephens J, Griksaitis M. Thoracic ultrasound: it's not all about the pleura. BMJ Open Respir Res. 2018;5:e000354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Amatya Y, Rupp J, Russell FM, Saunders J, Bales B, House DR. Diagnostic use of lung ultrasound compared to chest radiograph for suspected pneumonia in a resource-limited setting. Int J Emerg Med. 2018;11:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 17. | Gil-Rodríguez J, Pérez de Rojas J, Aranda-Laserna P, Benavente-Fernández A, Martos-Ruiz M, Peregrina-Rivas JA, Guirao-Arrabal E. Ultrasound findings of lung ultrasonography in COVID-19: A systematic review. Eur J Radiol. 2022;148:110156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 18. | Langlotz CP, Allen B, Erickson BJ, Kalpathy-Cramer J, Bigelow K, Cook TS, Flanders AE, Lungren MP, Mendelson DS, Rudie JD, Wang G, Kandarpa K. A Roadmap for Foundational Research on Artificial Intelligence in Medical Imaging: From the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology. 2019;291:781-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 210] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 19. | Abu Anas EM, Seitel A, Rasoulian A, St John P, Pichora D, Darras K, Wilson D, Lessoway VA, Hacihaliloglu I, Mousavi P, Rohling R, Abolmaesumi P. Bone enhancement in ultrasound using local spectrum variations for guiding percutaneous scaphoid fracture fixation procedures. Int J Comput Assist Radiol Surg. 2015;10:959-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60:84-90. [DOI] [Full Text] |
| 21. | Currie GM. Intelligent Imaging: Artificial Intelligence Augmented Nuclear Medicine. J Nucl Med Technol. 2019;47:217-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Liu S, Wang Y, Yang X, Lei B, Liu L, Li SX, Ni D, Wang T. Deep learning in medical ultrasound analysis: A review. Engineering 2019; 5: 261-275. [DOI] [Full Text] |
| 23. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36149] [Cited by in RCA: 21166] [Article Influence: 1924.2] [Reference Citation Analysis (2)] |
| 24. | Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18665] [Cited by in RCA: 18040] [Article Influence: 1061.2] [Reference Citation Analysis (1)] |
| 25. | Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13930] [Cited by in RCA: 13810] [Article Influence: 812.4] [Reference Citation Analysis (3)] |
| 26. | Arntfield R, VanBerlo B, Alaifan T, Phelps N, White M, Chaudhary R, Ho J, Wu D. Development of a convolutional neural network to differentiate among the etiology of similar appearing pathological B lines on lung ultrasound: a deep learning study. BMJ Open. 2021;11:e045120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (1)] |
| 27. | Awasthi N, Dayal A, Cenkeramaddi LR, Yalavarthy PK. Mini-COVIDNet: Efficient Lightweight Deep Neural Network for Ultrasound Based Point-of-Care Detection of COVID-19. IEEE Trans Ultrason Ferroelectr Freq Control. 2021;68:2023-2037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 28. | Barros B, Lacerda P, Albuquerque C, Conci A. Pulmonary COVID-19: Learning Spatiotemporal Features Combining CNN and LSTM Networks for Lung Ultrasound Video Classification. Sensors (Basel). 2021;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Born J, Wiedemann N, Cossio M, Buhre C, Brändle G, Leidermann K, Aujayeb A, Moor M, Rieck B, et al Accelerating detection of lung pathologies with explainable ultrasound image analysis. Appl Sci 2021; 11: 672. [RCA] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 30. | Born J, Brändle G, Cossio M, Disdier M, Goulet J, Roulin J, Wiedemann N. POCOVID-Net: Automatic detection of COVID-19 from a new lung ultrasound dataset (POCUS). ISMB TransMed COSI 2020 2021. Preprint. |
| 31. | Chen J, He C, Yin J, Li J, Duan X, Cao Y, Sun L, Hu M, Li W, Li Q. Quantitative Analysis and Automated Lung Ultrasound Scoring for Evaluating COVID-19 Pneumonia With Neural Networks. IEEE Trans Ultrason Ferroelectr Freq Control. 2021;68:2507-2515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 32. | Dastider AG, Sadik F, Fattah SA. An integrated autoencoder-based hybrid CNN-LSTM model for COVID-19 severity prediction from lung ultrasound. Comput Biol Med. 2021;132:104296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 33. | Diaz-Escobar J, Ordóñez-Guillén NE, Villarreal-Reyes S, Galaviz-Mosqueda A, Kober V, Rivera-Rodriguez R, Lozano Rizk JE. Deep-learning based detection of COVID-19 using lung ultrasound imagery. PLoS One. 2021;16:e0255886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 34. | Erfanian Ebadi S, Krishnaswamy D, Bolouri SES, Zonoobi D, Greiner R, Meuser-Herr N, Jaremko JL, Kapur J, Noga M, Punithakumar K. Automated detection of pneumonia in lung ultrasound using deep video classification for COVID-19. Inform Med Unlocked. 2021;25:100687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 35. | Hu Z, Liu Z, Dong Y, Liu J, Huang B, Liu A, Huang J, Pu X, Shi X, Yu J, Xiao Y, Zhang H, Zhou J. Evaluation of lung involvement in COVID-19 pneumonia based on ultrasound images. Biomed Eng Online. 2021;20:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 36. | La Salvia M, Secco G, Torti E, Florimbi G, Guido L, Lago P, Salinaro F, Perlini S, Leporati F. Deep learning and lung ultrasound for Covid-19 pneumonia detection and severity classification. Comput Biol Med. 2021;136:104742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 37. | Mento F, Perrone T, Fiengo A, Smargiassi A, Inchingolo R, Soldati G, Demi L. Deep learning applied to lung ultrasound videos for scoring COVID-19 patients: A multicenter study. J Acoust Soc Am. 2021;149:3626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 38. | Roy S, Menapace W, Oei S, Luijten B, Fini E, Saltori C, Huijben I, Chennakeshava N, Mento F, Sentelli A, Peschiera E, Trevisan R, Maschietto G, Torri E, Inchingolo R, Smargiassi A, Soldati G, Rota P, Passerini A, van Sloun RJG, Ricci E, Demi L. Deep Learning for Classification and Localization of COVID-19 Markers in Point-of-Care Lung Ultrasound. IEEE Trans Med Imaging. 2020;39:2676-2687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 237] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 39. | Sadik F, Dastider AG, Fattah SA. SpecMEn-DL: spectral mask enhancement with deep learning models to predict COVID-19 from lung ultrasound videos. Health Inf Sci Syst. 2021;9:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 40. | Muhammad G, Shamim Hossain M. COVID-19 and Non-COVID-19 Classification using Multi-layers Fusion From Lung Ultrasound Images. Inf Fusion. 2021;72:80-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 41. | Tsai CH, van der Burgt J, Vukovic D, Kaur N, Demi L, Canty D, Wang A, Royse A, Royse C, Haji K, Dowling J, Chetty G, Fontanarosa D. Automatic deep learning-based pleural effusion classification in lung ultrasound images for respiratory pathology diagnosis. Phys Med. 2021;83:38-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 42. | Xue W, Cao C, Liu J, Duan Y, Cao H, Wang J, Tao X, Chen Z, Wu M, Zhang J, Sun H, Jin Y, Yang X, Huang R, Xiang F, Song Y, You M, Zhang W, Jiang L, Zhang Z, Kong S, Tian Y, Zhang L, Ni D, Xie M. Modality alignment contrastive learning for severity assessment of COVID-19 from lung ultrasound and clinical information. Med Image Anal. 2021;69:101975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 43. |
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D.
Grad-cam: Visual explanations from deep networks |
| 44. | Soldati G, Smargiassi A, Inchingolo R, Buonsenso D, Perrone T, Briganti DF, Perlini S, Torri E, Mariani A, Mossolani EE, Tursi F, Mento F, Demi L. Proposal for International Standardization of the Use of Lung Ultrasound for Patients With COVID-19: A Simple, Quantitative, Reproducible Method. J Ultrasound Med. 2020;39:1413-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 434] [Article Influence: 72.3] [Reference Citation Analysis (1)] |
| 45. | Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, Adam M. Mobilenets: Efficient convolutional neural networks for mobile vision applications; 2017. Preprint. Cited 17 April 2017. |
| 46. | Zhou Z, Sodha V, Rahman Siddiquee MM, Feng R, Tajbakhsh N, Gotway MB, Liang J. Models genesis: Generic autodidactic models for 3D medical image analysis. In Shen D, Liu T, Peters TM, Staib LH, Essert C, Zhou S, Yap PT, Khan A editors. Medical Image Computing and Computer Assisted Intervention–MICCAI 2019. Proceedings of 22nd International Conference MICCAI; 2019 Oct 13-17; Shenzhen, China. Lecture Notes in Computer Science. Springer, Cham, Switzerland, 2019; 11767: 384-393. [RCA] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 47. | Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ. Advances in neural information processing systems. New York: Curran Associates Inc, 2012; 25: 1097–1105. |
| 48. | Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, et al ImageNet large scale visual recognition challenge. Int J Comput Vis 2015; 115: 211–252. [RCA] [DOI] [Full Text] [Cited by in Crossref: 16940] [Cited by in RCA: 6807] [Article Influence: 618.8] [Reference Citation Analysis (0)] |
| 49. | Horry MJ, Chakraborty S, Paul M, Ulhaq A, Pradhan B, Saha M, Shukla N. Covid-19 detection through transfer learning using multimodal imaging data. IEEE Access 2020; 8: 149808–149824. [RCA] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 168] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 50. | Moran M, Faria M, Giraldi G, Bastos L, Oliveira L, Conci A. Classification of Approximal Caries in Bitewing Radiographs Using Convolutional Neural Networks. Sensors (Basel). 2021;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 51. | LeCun Y, Haffner P, Bottou L, Bengio Y. Object recognition with gradient-based learning. In: Shape, Contour and Grouping in Computer Vision. Lecture Notes in Computer Science, vol 1681. Springer: Berlin/Heidelberg, Germany, 1999. [RCA] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 108] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Radiology, nuclear medicine and medical imaging
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Haurylenka D, Belarus; Zhang W, China S-Editor: Liu JH L-Editor: A P-Editor: Liu JH