Published online Jun 28, 2021. doi: 10.35711/aimi.v2.i3.64
Peer-review started: May 13, 2021
First decision: June 2, 2021
Revised: June 19, 2021
Accepted: June 30, 2021
Article in press: June 30, 2021
Published online: June 28, 2021
Processing time: 57 Days and 1.9 Hours
Hepatocellular carcinoma (HCC) is the most common form of primary liver cancer with low 5-year survival rate. The high molecular heterogeneity in HCC poses huge challenges for clinical practice or trial design and has become a major barrier to improving the management of HCC. However, current clinical practice based on single bioptic or archived tumor tissue has been deficient in identifying useful biomarkers. The concept of radiomics was first proposed in 2012 and is different from the traditional imaging analysis based on the qualitative or semi-quantitative analysis by radiologists. Radiomics refers to high-throughput extraction of large amounts number of high-dimensional quantitative features from medical images through machine learning or deep learning algorithms. Using the radiomics method could quantify tumoral phenotypes and heterogeneity, which may provide benefits in clinical decision-making at a lower cost. Here, we review the workflow and application of radiomics in HCC.
Core Tip: The high molecular heterogeneity in hepatocellular carcinoma poses huge challenges for clinical practice or trial design and has become a major barrier to improving the management of hepatocellular carcinoma. Radiomics could quantify tumoral phenotypes and heterogeneity, which may provide benefits in clinical decision-making at a lower cost. Here, we review the workflow and application of radiomics in hepatocellular carcinoma.
- Citation: Jin ZC, Zhong BY. Application of radiomics in hepatocellular carcinoma: A review. Artif Intell Med Imaging 2021; 2(3): 64-72
- URL: https://www.wjgnet.com/2644-3260/full/v2/i3/64.htm
- DOI: https://dx.doi.org/10.35711/aimi.v2.i3.64
Liver cancer is one of the most common malignant tumors worldwide. There are approximately 906000 new cases and 830000 deaths every year, ranking as the sixth most commonly diagnosed cancer and the third mortality[1]. Hepatocellular carcinoma (HCC) comprises 75%-85% of cases of primary liver cancer. There is high molecular heterogeneity in HCC at three levels, including the heterogeneity between tumor nodules within the same individual (intertumoral heterogeneity), between different regions of the same tumor nodule (intratumor heterogeneity), and between patients (interpatient heterogeneity)[2]. HCC has one of the fewest somatic mutations in solid tumors that can be targeted by molecular therapies and of which treatment response could not be predicted by mutations in clinical practice[3]. These characteristics of HCC pose huge challenges for clinical practice or trial design and have become a major barrier to improving the management of HCC[4,5]. However, current clinical practice based on single bioptic or archived tumor tissue has been deficient in identifying useful biomarkers[5].
Radiomics was first proposed in 2012 and is different from the traditional imaging analysis based on the qualitative or semi-quantitative analysis by radiologists[6]. This method refers to high-throughput extraction of large amounts of high-dimensional quantitative features from medical images through machine learning (ML) or deep learning (DL) algorithms[7,8]. These features that have been transformed into minable data could be used for diagnosis, treatment evaluation, and prognosis prediction[9]. Using the radiomics method could quantify tumoral phenotypes and heterogeneity, which may provide benefits in clinical decision-making at a lower cost[10,11]. Here, we review the workflow and application of radiomics in HCC.
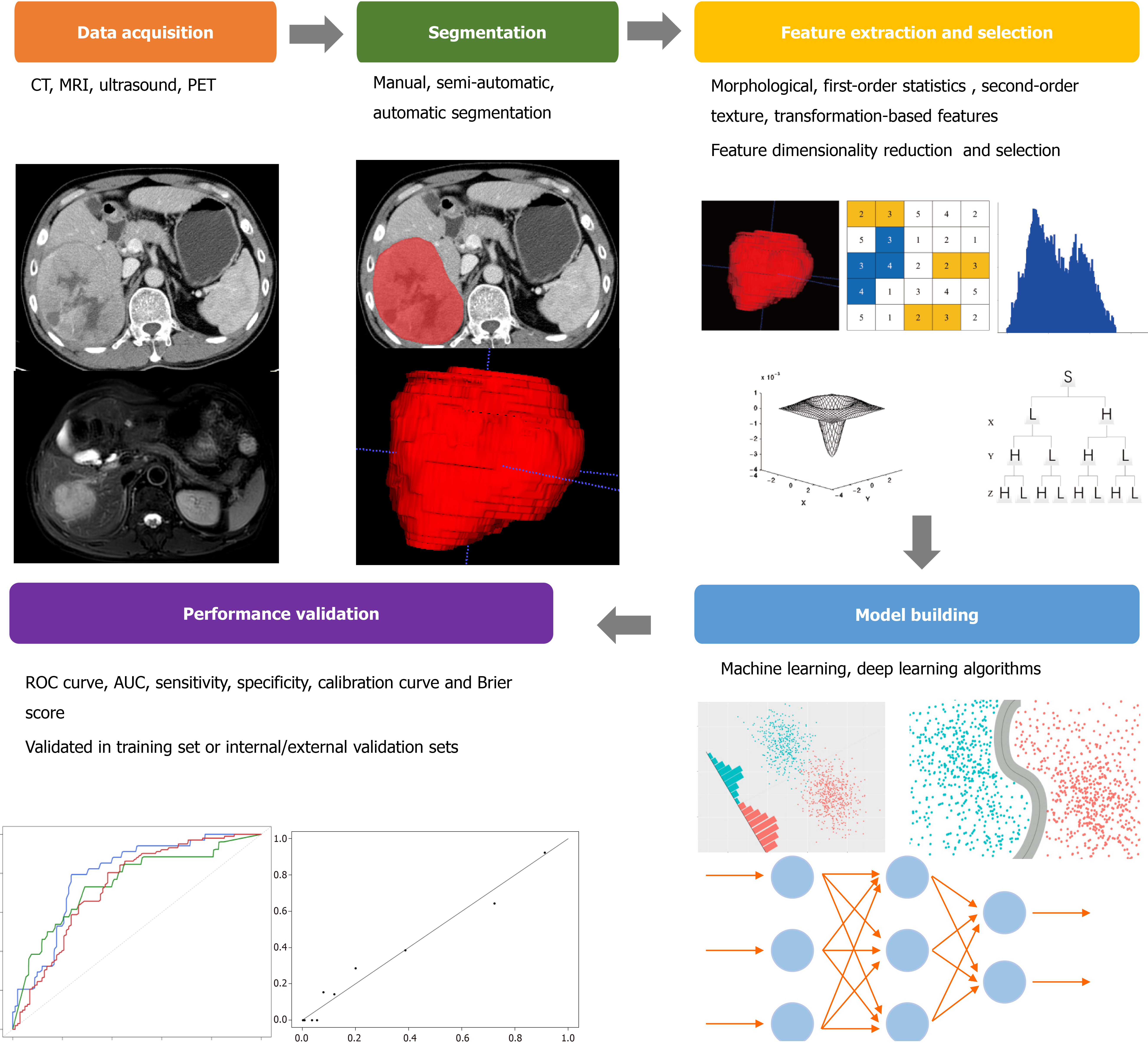
The workflow of radiomics mainly includes: image data acquisition and preprocessing, the volume of interest (VOI) segmentation, feature extraction, model establishment, and performance validation (Figure 1)[9].
Although radiomics was first and widely utilized in computed tomography (CT) and magnetic resonance imaging (MRI) images, there were more and more studies using ultrasound (US) as well as positron emission tomography images. Most studies were conducted based on retrospective image data sets, even different hospitals and different scanning equipment. The standardized imaging protocols could reduce the unnecessary confounding variability, or it will affect the quality and stability of the extracted imaging features. A previous study found that the feature variability caused by different CT scanners was even comparable to the feature variability found in the tumor[12]. The disclosed imaging protocols were suggested to increase the reproducibility and comparability in future radiomics studies[9].
The three-dimensional VOI segmentation that captures the tumor comprehensive panorama could be delineated by using manual, semi-automatic, and automatic segmentation methods. However, the variability in the segmentation process inevitably introduces bias. Meanwhile, the partial volume effect makes the segmentation challenge that could lead to the blurring of the edge and morphological variation of the lesion. Multiple segmentation is an effective method that can limit bias and help to select robust features, including the evaluation by multiple clinicians and the combination of different segmentation algorithms. However, the commonly used segmentation method in radiomics is manual segmentation and relies on an experienced clinician, which is quite boring and time-consuming. Several semi-automatic or automatic segmentation methods have been reported[13,14]. These methods could minimize labor costs and improve the repeatability and reliability of studies but are not widely recognized and applied.
The high-throughput extraction of quantitative features from VOI is the key process in radiomics analysis after appropriate image preprocessing. The imaging features that are empirically defined by radiologists are named semantic features. These features cannot be described by specific mathematical expressions nor can they be specifically extracted from images, but they are still meaningful in imaging interpretation and clinical application. These non-semantic features quantitatively described by mathematical expressions can usually be divided into four categories: morphological features, first-order statistics features, second-order texture features, and transformation-based features. Morphological features describe the three-dimensional and two- dimensional size and shape of VOI, such as diameter, perimeter, sphericity, and flatness. First-order statistics features (also called histogram features) evaluate the gray-level frequency distribution in VOI, including maximum, median, minimum, and entropy, while second-order texture features are often derived from the gray-level matrix and describe the statistical relationship between voxel gray levels, including gray-level co-occurrence matrix and gray level run length matrix. The voxel gray-level patterns in different spatial frequencies are analyzed by transformation-based features, including Fourier, Gabor, and wavelet features.
According to the number of filters, feature categories, and other parameters, the number of features extracted from the images can be infinite. The inclusion of all relevant features in a predictive model inevitably leads to overfitting, which negatively impacts the efficacy of its prediction performance. It is necessary to introduce a feature selection method to eliminate unsuitable features, that is, feature dimensionality reduction methods (such as principal component analysis or clustering). By reducing redundant and interference items by dimensionality reduction, the features for further analysis contain useful and repeatable information to a large extent.
The prediction model composed of selected features was constructed by an ML algorithm, including support vector machine, random forest, linear discriminant analysis, and so on. The specific method was chosen according to the preference and experience of the researchers. However, different modeling methods have been proved to affect the prediction performance of imaging models and have inherent limitations, such as the independence assumption in logistic regression, feature discretization in Bayesian networks, or network structure dependence in DL. Therefore, a variety of ways could be considered to build the model in the study.
The predictive performance evaluation of the model requires an internal or external validation set to determine whether the model has good generalization performance or only predictability for the specific samples analyzed. This process is often measured by the receiver operating characteristic curve with area under the curve (AUC), sensitivity, and specificity. In addition, the consistency between the observed results and the model prediction was also evaluated necessarily, which can be evaluated by the calibration curve and Brier score. An effective model shows consistency in both training and validation sets. The models validated by an independent external set are more reliable than those validated by an internal set, and of course, the models that could be prospectively verified are more persuasive.
Imaging is a crucial part of the HCC diagnosis. Multiphasic contrast-enhanced CT or contrast-enhanced MRI should be used first with high sensitivity recommended by the European Association for the Study of the Liver[15]. Li et al[16] extracted the texture features from the SPAIR T2WI sequence in MRI and used four different classifiers to identify single intrahepatic lesions (hepatic hemangioma, hepatic metastases, and HCC). The error rates were 11.7% (hepatic hemangioma vs hepatic metastases), 9.6% (hepatic metastases vs HCC), and 9.7% (hepatic hemangioma vs HCC). The combination of quantitative apparent diffusion coefficient histogram parameters and the Liver Imaging Reporting And Data System could distinguish HCC from other subtypes of primary liver cancer, such as intrahepatic cholangiocarcinoma and mixed HCC-intrahepatic cholangiocarcinoma[17]. A total of 63 patients confirmed by pathology were included, and it was found that the model combined with gender, Liver Imaging Reporting And Data System, and the fifth percentile apparent diffusion coefficient could achieve a good prediction efficiency. The AUCs could reach 0.90/0.89 with the accuracy of 81.5%/80.0%, the sensitivity of 79.3%/86.2%, and the specificity of 88.9%/77.8% for two independent observers. Huang et al[18] managed to distinguish dual phenotypic HCC by different classifiers based on Gd-EOB-DTPA-enhanced MRI and showed good predictive performance.
For the new HCC nodules in patients with a liver cirrhosis background, radiomics features extracted from multiphasic contrast-enhanced CT combined with the ML algorithm could bring benefits. Mokrane et al[19] retrospectively included 178 patients from 27 centers and divided them into a training set (142 patients) and validation set (36 patients). All the patients had nodules that were classified as indeterminate liver nodules by the European Association for the Study of the Liver guidelines, and the histological classification was finally confirmed by liver biopsy. A total of 13920 quantitative radiomics features were extracted from the plain, arterial, venous, and dual-phase (delta) phases. Three supervised ML classification algorithms: K nearest neighbor, support vector machine, and random forest algorithm were used to establish the models. A single feature was finally obtained, which represented the characteristics of changes in nodule phenotype between arterial and portal venous phases (corresponds to the “washout” pattern during the contrast agent clearance). Finally, the radiomics signature used reached an AUC value of 0.66 with a sensitivity of 0.70 and specificity of 0.59 in the external validation set.
US is one of the important methods in the diagnostic algorithm and recall policy by the European Association for the Study of the Liver guidelines[15]. However, US images are more heterogeneous because of the images acquired by different clinicians with multiple examination parameters. There was a study that reported that the features extracted from US images could be classified by using neural network classifiers to distinguish focal liver lesions, including typical and atypical cysts, hepatic hemangiomas, liver metastases, and HCC lesions, with an accuracy of up to 95%[20]. A multitask DL algorithm was constructed that detects and characterizes focal liver lesions in a public dataset[21]. The model simultaneously yielded AUCs of 0.935 for lesion detection and 0.916 for focal liver lesions characterization (benign vs malignant).
Radiomics could effectively diagnose and distinguish the HCC lesion from the different intrahepatic lesions, new nodules, and even the subtypes of primary liver cancer. Although the above studies are based on different imaging modalities and ML/DL methods, this method is expected to further assist doctors in clinical diagnosis and decision-making in the future.
Surgical resection is the first choice for HCC patients with good performance status and liver function reserve. But the postoperative 5-year recurrence rate could be as high as 70%. To solve this problem, a multicenter retrospective study was carried out from three independent centers. The study included 295 early-stage HCC patients within Milan criteria who have received preoperative contrast-enhanced CT examination. Recurrence-free survival was selected as the primary endpoint of this study. Based on 177 patients from one center (training set), two prediction models have been constructed that incorporated preoperative variables or postoperative variables. The results showed that the prediction efficiency of the two radiomics-based models was higher than that of previous clinical models and staging systems and can well stratify patients with a low, moderate, and high risk of recurrence.
The application of radiomics in predicting postoperative recurrence has also been verified in other studies. In addition, some studies have found that the radiomics model based on preoperative MRI images can better predict the 5-year survival of patients after hepatectomy. Cai et al[22] retrospectively included 112 patients who underwent hepatectomy to predict postoperative liver failure by a radiomics-based nomogram. The AUC value of the training set was 0.822 (95% confidence interval: 0.753-0.917), and the AUC value of the validation set was 0.762 (95% confidence interval: 0.576-0.948). When it was compared with MELD, Child-Pugh, and ALBI score, the radiomics model showed a significant advantage. The researchers conducted a prospective validation analysis of 13 patients who underwent hepatectomy with an AUC of 0.833 (95% confidence interval: 0.591-1.000). Decision curve analysis showed that the model could bring clinical benefits. Radiomic features could identify the tumor invasion and predict recurrence after liver transplantation[23].
Ablation is recommended for HCC patients with Barcelona Clinic Liver Cancer 0 or A stage who are not suitable for surgery. Radiomic features extracted from perioperative CT images could predict early recurrence after curative ablation[24,25]. Among them, the features based on portal vein phase CT images performed best in the validation set. When the clinicopathological factors were added to the model, the portal vein phase-based combined model showed good prediction performance in the training/validation set and significantly better than that of the simple clinical model. Microwave ablation was performed in pigs under CT guidance for improving the visualization of post ablational coagulation necrosis in a proof of concept study[26]. The results showed that radiomic profiles of the fully necrotic areas seemed to be different from those areas with vital tissue. The subregion radiomics analysis could identify these differences with classification algorithms.
Transarterial chemoembolization (TACE) is the most widely used treatment for unresectable HCC in clinical practice. Radiomics plays a role in the prediction of treatment response to TACE[27-29]. Chen et al[27] analyzed the radiomic features extracted from tumoral VOI and peritumoral VOI, drawn at the hepatic arterial and non-contrast phases, respectively. The radiomic signature extracted from the peritumoral VOI with expanded 10 mm rim away from the main tumor part achieved excellent performance in predicting the first TACE response. Several studies established a radiomic model based on the preoperative images to predict long-term outcomes of patients who underwent TACE with good performance[30,31]. However, there were various confounding factors during multiple TACE sessions that may weaken the actual predictive performance. Fu et al[32] included 520 patients from five independent centers (divided into a training set and validation set). A comprehensive model including treatment (liver resection or TACE), age, sex, modified Barcelona Clinic Liver Cancer stage, fusion focus, tumor capsule, and three radiomic features was established with good differentiation and calibration. The AUC value of the predicted 3-year recurrence-free survival was 0.80 in the training set and 0.75 in the test set.
Sorafenib is the first oral multikinase inhibitor recommended in patients with advanced HCC. Various clinical trials tried to explore the possibility of combining sorafenib and TACE that may inhibit revascularization and tumor proliferation after TACE. Most of these trials failed, except the TACTICS trial conducted recently. It is important to identify HCC patients who may benefit from the combination of TACE plus sorafenib. A DL-based radiomic model provided a significant prediction value with an AUC value of 0.717 in the training set and 0.714 in the validation set[33].
Microvascular invasion (MVI) of HCC mainly refers to the presence of cancer cells in the endothelial-lined vascular lumen under the microscope, which is a powerful validated, important independent risk factor for early recurrence and poor survival after surgical resection of HCC. Radiomic features extracted from preoperative enhanced MRI multi-phase images could predict the occurrence of MVI favorably[34,35]. By using the least absolute shrinkage and selection operator method to select appropriate radiomic features, the predictive performance of the combined model incorporating clinicoradiological predictors and radiomic features was better than the clinicoradiological model (AUC 0.943 vs 0.850 in the training set, and 0.861 vs 0.759 in the validation set). The sensitivity, specificity, and accuracy of the combined model were 88.2%/89.5%, 87.5%/81.4%, and 87.7%/83.9% in two sets, respectively. Several studies reported that using contrast-enhanced CT images to develop and validate radiomics nomogram was a clinically useful tool to identify patients[36,37]. However, a retrospective study that included 495 patients with postoperative MVI status confirmed by histology (MVI- group, n = 346, and MVI + group, n = 149)[38] found that radiomics analysis with current CT imaging protocols does not provide significant additional value to the conventional semantic features.
Pathological grading of HCC is one of the factors that influence prognosis. Most patients with high-grade tumors have a higher rate of intrahepatic recurrence than those without low-grade tumors. The radiomics signatures based on MRI T1WI or T2WI images could be helpful for the preoperative prediction of the pathological grade of HCC[39]. The combination of the radiomic signatures and clinical factors achieved the best predictive performance over the other simple model and distinguished between high-grade and low-grade HCC (AUC = 0.800). In addition, cytokeratin 19 status of HCC that is associated with clinical aggressiveness could be identified by a radiomic-based model with satisfactory prediction performance[40]. Ye et al[41] managed to use the texture feature analysis on gadoxetic acid-enhanced MRI images preoperatively to predict Ki-67 status of HCC. However, the optimal cut-off value of the Ki-67 level was defined by the researcher, which weakened the generalization of the study.
Gene expression patterns of cancer tissues could reflect the underlying cellular pathophysiology and enrich the understanding of cellular pathways and numerous pathological conditions. Imaging traits have the potential to be a surrogate marker of the clinically relevant genomic/molecular signature of HCC[42-45]. One study found that the dynamic imaging traits from CT systematically correlated with the global gene expression programs of HCC[42]. The combination of 28 imaging traits was sufficient to reconstruct the variation of 116 gene expression profiles, revealing cell proliferation, liver synthetic function, and patient prognosis. Moreover, they developed a two-imaging-trait decision tree, including internal arteries and hypodense halos in HCC that is associated with a gene expression signature of venous invasion and could predict histologic venous invasion and survival of patients. Based on that result, a similar team defined a contrast-enhanced CT imaging biomarker for predicting MVI named radiogenomic venous invasion[43]. In a multicenter retrospective study, the radiogenomic venous invasion biomarker was a robust predictor of MVI with a diagnostic accuracy of 89%, sensitivity of 76%, and specificity of 94% and was associated with a poor overall survival that could have broad clinical use. They considered that radiogenomic venous invasion derived from a gene expression signature of venous invasion may reflect a more fundamental phenotype of the tumor.
Qualitative and quantitative MRI radiomic features could serve as the noninvasive biomarker to predict HCC immuno-oncological characteristics and tumor recurrence[46]. One study analyzed the correlation between radiomics, immunoprofiling (CD3, CD68, CD31), and genomic (PD-1 at the protein level, PD-L1 and CTLA4 at the mRNA expression level) features with statistical significance[46]. Radiomic features, including tumor size, showed good prediction performance for early HCC recurrence after resection, while immunoprofiling and genomic features did not.
Systemic therapy in advanced HCC has developed rapidly in recent years, with the most prominent success of the combination of atezolizumab (anti-PD-L1 antibody) and bevacizumab (anti-VEGF antibody). However, due to the huge tumor heterogeneity in HCC, several promising trials (such as keynote-240 and checkmate-459) have failed, and the best objective response rates of successful systemic therapies are only around 30%. In addition, there are more and more ongoing trials in the adjuvant or combination therapies setting of HCC that are explored and practiced currently. Personalized treatment and more precise patient stratification may be required under such circumstances. Radiomics technology based on ML/DL algorithms is expected to become a bridge that connects the clinical personalized precision treatment of HCC patients and its tumor phenotype. Further radiomics research with multicenter and prospective validation is still needed for improving its interpretability and reproducibility.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68648] [Article Influence: 13729.6] [Reference Citation Analysis (201)] |
| 2. | Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:139-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 597] [Article Influence: 99.5] [Reference Citation Analysis (1)] |
| 3. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2066] [Cited by in RCA: 3369] [Article Influence: 481.3] [Reference Citation Analysis (45)] |
| 4. | Lin DC, Mayakonda A, Dinh HQ, Huang P, Lin L, Liu X, Ding LW, Wang J, Berman BP, Song EW, Yin D, Koeffler HP. Genomic and Epigenomic Heterogeneity of Hepatocellular Carcinoma. Cancer Res. 2017;77:2255-2265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 161] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 5. | Lu LC, Hsu CH, Hsu C, Cheng AL. Tumor Heterogeneity in Hepatocellular Carcinoma: Facing the Challenges. Liver Cancer. 2016;5:128-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 6. | Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2415] [Cited by in RCA: 4195] [Article Influence: 299.6] [Reference Citation Analysis (4)] |
| 7. | Piccialli F, Calabrò F, Crisci D, Cuomo S, Prezioso E, Mandile R, Troncone R, Greco L, Auricchio R. Precision medicine and machine learning towards the prediction of the outcome of potential celiac disease. Sci Rep. 2021;11:5683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 8. | Piccialli F, Somma VD, Giampaolo F, Cuomo S, Fortino G. A survey on deep learning in medicine: Why, how and when? Information Fusion. 2021;66:111-137. [DOI] [Full Text] |
| 9. | Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1825] [Cited by in RCA: 3951] [Article Influence: 439.0] [Reference Citation Analysis (0)] |
| 10. | Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2262] [Cited by in RCA: 3459] [Article Influence: 288.3] [Reference Citation Analysis (0)] |
| 11. | O'Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21:249-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 512] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 12. | Mackin D, Fave X, Zhang LF, Fried D, Yang JZ, Taylor B, Rodriguez-Rivera E, Dodge C, Jones AK, Court L. Measuring Computed Tomography Scanner Variability of Radiomics Features. Invest Radiol. 2015;50:757-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 509] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 13. | Heye T, Merkle EM, Reiner CS, Davenport MS, Horvath JJ, Feuerlein S, Breault SR, Gall P, Bashir MR, Dale BM, Kiraly AP, Boll DT. Reproducibility of dynamic contrast-enhanced MR imaging. Part II. Comparison of intra- and interobserver variability with manual region of interest placement versus semiautomatic lesion segmentation and histogram analysis. Radiology. 2013;266:812-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 14. | Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, Mitra S, Shankar BU, Kikinis R, Haibe-Kains B, Lambin P, Aerts HJ. Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One. 2014;9:e102107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 354] [Cited by in RCA: 441] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 15. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5593] [Cited by in RCA: 6422] [Article Influence: 802.8] [Reference Citation Analysis (9)] |
| 16. | Li Z, Mao Y, Huang W, Li H, Zhu J, Li W, Li B. Texture-based classification of different single liver lesion based on SPAIR T2W MRI images. BMC Med Imaging. 2017;17:42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 17. | Lewis S, Peti S, Hectors SJ, King M, Rosen A, Kamath A, Putra J, Thung S, Taouli B. Volumetric quantitative histogram analysis using diffusion-weighted magnetic resonance imaging to differentiate HCC from other primary liver cancers. Abdom Radiol (NY). 2019;44:912-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 18. | Huang X, Long L, Wei J, Li Y, Xia Y, Zuo P, Chai X. Radiomics for diagnosis of dual-phenotype hepatocellular carcinoma using Gd-EOB-DTPA-enhanced MRI and patient prognosis. J Cancer Res Clin Oncol. 2019;145:2995-3003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 19. | Mokrane FZ, Lu L, Vavasseur A, Otal P, Peron JM, Luk L, Yang H, Ammari S, Saenger Y, Rousseau H, Zhao B, Schwartz LH, Dercle L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30:558-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 20. | Virmani J, Kumar V, Kalra N, Khandelwal N. Neural network ensemble based CAD system for focal liver lesions from B-mode ultrasound. J Digit Imaging. 2014;27:520-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aubé C, Luciani A, Lassau N, Jégou S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;100:227-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 22. | Cai W, He B, Hu M, Zhang W, Xiao D, Yu H, Song Q, Xiang N, Yang J, He S, Huang Y, Huang W, Jia F, Fang C. A radiomics-based nomogram for the preoperative prediction of posthepatectomy liver failure in patients with hepatocellular carcinoma. Surg Oncol. 2019;28:78-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 23. | Guo D, Gu D, Wang H, Wei J, Wang Z, Hao X, Ji Q, Cao S, Song Z, Jiang J, Shen Z, Tian J, Zheng H. Radiomics analysis enables recurrence prediction for hepatocellular carcinoma after liver transplantation. Eur J Radiol. 2019;117:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (1)] |
| 24. | Yuan C, Wang Z, Gu D, Tian J, Zhao P, Wei J, Yang X, Hao X, Dong D, He N, Sun Y, Gao W, Feng J. Prediction early recurrence of hepatocellular carcinoma eligible for curative ablation using a Radiomics nomogram. Cancer Imaging. 2019;19:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 25. | Shan QY, Hu HT, Feng ST, Peng ZP, Chen SL, Zhou Q, Li X, Xie XY, Lu MD, Wang W, Kuang M. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging. 2019;19:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 142] [Article Influence: 20.3] [Reference Citation Analysis (1)] |
| 26. | Bressem KK, Adams LC, Vahldiek JL, Erxleben C, Poch F, Lehmann KS, Hamm B, Niehues SM. Subregion Radiomics Analysis to Display Necrosis After Hepatic Microwave Ablation-A Proof of Concept Study. Invest Radiol. 2020;55:422-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 27. | Chen M, Cao J, Hu J, Topatana W, Li S, Juengpanich S, Lin J, Tong C, Shen J, Zhang B, Wu J, Pocha C, Kudo M, Amedei A, Trevisani F, Sung PS, Zaydfudim VM, Kanda T, Cai X. Clinical-Radiomic Analysis for Pretreatment Prediction of Objective Response to First Transarterial Chemoembolization in Hepatocellular Carcinoma. Liver Cancer. 2021;10:38-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 101] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 28. | Park HJ, Kim JH, Choi SY, Lee ES, Park SJ, Byun JY, Choi BI. Prediction of Therapeutic Response of Hepatocellular Carcinoma to Transcatheter Arterial Chemoembolization Based on Pretherapeutic Dynamic CT and Textural Findings. AJR Am J Roentgenol. 2017;209:W211-W220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 29. | Jin Z, Chen L, Zhong B, Zhou H, Zhu H, Song J, Guo J, Zhu X, Ji J, Ni C, Teng G. Machine-learning analysis of contrast-enhanced computed tomography radiomics predicts patients with hepatocellular carcinoma who are unsuitable for initial transarterial chemoembolization monotherapy: A multicenter study. Transl Oncol. 2021;14:101034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 30. | Kong C, Zhao Z, Chen W, Lv X, Shu G, Ye M, Song J, Ying X, Weng Q, Weng W, Fang S, Chen M, Tu J, Ji J. Prediction of tumor response via a pretreatment MRI radiomics-based nomogram in HCC treated with TACE. Eur Radiol. 2021 epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 31. | Zhao Y, Wang N, Wu J, Zhang Q, Lin T, Yao Y, Chen Z, Wang M, Sheng L, Liu J, Song Q, Wang F, An X, Guo Y, Li X, Wu T, Liu AL. Radiomics Analysis Based on Contrast-Enhanced MRI for Prediction of Therapeutic Response to Transarterial Chemoembolization in Hepatocellular Carcinoma. Front Oncol. 2021;11:582788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 32. | Fu S, Wei J, Zhang J, Dong D, Song J, Li Y, Duan C, Zhang S, Li X, Gu D, Chen X, Hao X, He X, Yan J, Liu Z, Tian J, Lu L. Selection Between Liver Resection Versus Transarterial Chemoembolization in Hepatocellular Carcinoma: A Multicenter Study. Clin Transl Gastroenterol. 2019;10:e00070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Zhang L, Xia W, Yan ZP, Sun JH, Zhong BY, Hou ZH, Yang MJ, Zhou GH, Wang WS, Zhao XY, Jian JM, Huang P, Zhang R, Zhang S, Zhang JY, Li Z, Zhu XL, Gao X, Ni CF. Deep Learning Predicts Overall Survival of Patients With Unresectable Hepatocellular Carcinoma Treated by Transarterial Chemoembolization Plus Sorafenib. Front Oncol. 2020;10:593292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 34. | Yang L, Gu D, Wei J, Yang C, Rao S, Wang W, Chen C, Ding Y, Tian J, Zeng M. A Radiomics Nomogram for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma. Liver Cancer. 2019;8:373-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 259] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 35. | Song D, Wang Y, Wang W, Cai J, Zhu K, Lv M, Gao Q, Zhou J, Fan J, Rao S, Wang M, Wang X. Using deep learning to predict microvascular invasion in hepatocellular carcinoma based on dynamic contrast-enhanced MRI combined with clinical parameters. J Cancer Res Clin Oncol. 2021 epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 36. | Ma X, Wei J, Gu D, Zhu Y, Feng B, Liang M, Wang S, Zhao X, Tian J. Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol. 2019;29:3595-3605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 177] [Article Influence: 25.3] [Reference Citation Analysis (1)] |
| 37. | Ni M, Zhou X, Lv Q, Li Z, Gao Y, Tan Y, Liu J, Liu F, Yu H, Jiao L, Wang G. Radiomics models for diagnosing microvascular invasion in hepatocellular carcinoma: which model is the best model? Cancer Imaging. 2019;19:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (1)] |
| 38. | Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, Yang G, Yan X, Zhang YD, Liu XS. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70:1133-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 546] [Article Influence: 78.0] [Reference Citation Analysis (1)] |
| 39. | Wu M, Tan H, Gao F, Hai J, Ning P, Chen J, Zhu S, Wang M, Dou S, Shi D. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol. 2019;29:2802-2811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 140] [Article Influence: 17.5] [Reference Citation Analysis (1)] |
| 40. | Wang W, Gu D, Wei J, Ding Y, Yang L, Zhu K, Luo R, Rao SX, Tian J, Zeng M. A radiomics-based biomarker for cytokeratin 19 status of hepatocellular carcinoma with gadoxetic acid-enhanced MRI. Eur Radiol. 2020;30:3004-3014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 41. | Ye Z, Jiang H, Chen J, Liu X, Wei Y, Xia C, Duan T, Cao L, Zhang Z, Song B. Texture analysis on gadoxetic acid enhanced-MRI for predicting Ki-67 status in hepatocellular carcinoma: A prospective study. Chin J Cancer Res. 2019;31:806-817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 42. | Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY, Kuo MD. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 450] [Article Influence: 23.7] [Reference Citation Analysis (2)] |
| 43. | Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, Rutman AM, Siripongsakun S, Lu D, Imanbayev G, Kuo MD. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62:792-800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 292] [Article Influence: 26.5] [Reference Citation Analysis (1)] |
| 44. | Taouli B, Hoshida Y, Kakite S, Chen X, Tan PS, Sun X, Kihira S, Kojima K, Toffanin S, Fiel MI, Hirschfield H, Wagner M, Llovet JM. Imaging-based surrogate markers of transcriptome subclasses and signatures in hepatocellular carcinoma: preliminary results. Eur Radiol. 2017;27:4472-4481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 45. | Xia W, Chen Y, Zhang R, Yan Z, Zhou X, Zhang B, Gao X. Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data-a preliminary study. Phys Med Biol. 2018;63:035044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 46. | Hectors SJ, Lewis S, Besa C, King MJ, Said D, Putra J, Ward S, Higashi T, Thung S, Yao S, Laface I, Schwartz M, Gnjatic S, Merad M, Hoshida Y, Taouli B. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur Radiol. 2020;30:3759-3769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 121] [Article Influence: 20.2] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Calabro F S-Editor: Liu M L-Editor: Filipodia P-Editor: Xing YX