Published online Jun 8, 2025. doi: 10.35712/aig.v6.i1.105682
Revised: April 5, 2025
Accepted: May 24, 2025
Published online: June 8, 2025
Processing time: 123 Days and 17.3 Hours
Celiac disease (CD) is a common autoimmune disorder where gluten ingestion triggers an immune response, damaging the small intestine in genetically pre
Core Tip: Advances in precision medicine, powered by artificial intelligence (AI), machine learning (ML) and deep learning (DL), are transforming the diagnosis and management of celiac disease (CD). By integrating histopathologic features, genetic data, and electronic medical records (EMRs), these technologies enable accurate disease classification, risk prediction, and personalized treatment strategies. ML and DL models have demonstrated high accuracy in predicting CD severity, while EMR-based approaches facilitate tailored dietary and therapeutic interventions. These innovations underscore the potential of AI-driven methodologies to enhance patient outcomes and revolutionize the clinical approach to CD.
- Citation: Rahmoune H, Boutrid N, Benchoufi I. Precision medicine in celiac disease: A step ahead. Artif Intell Gastroenterol 2025; 6(1): 105682
- URL: https://www.wjgnet.com/2644-3236/full/v6/i1/105682.htm
- DOI: https://dx.doi.org/10.35712/aig.v6.i1.105682
Celiac disease (CD) is a chronic immune-mediated enteropathy triggered by the ingestion of gluten—a protein found in wheat, barley, and rye—in genetically predisposed individuals[1]. It is estimated to affect approximately 1% of the global population, making it one of the most common autoimmune disorders worldwide[2,3].
The clinical manifestations of CD are highly variable and can be classified into gastrointestinal and extraintestinal symptoms. Gastrointestinal symptoms include diarrhea, abdominal pain, and bloating, while extraintestinal manifestations encompass conditions like anemia, osteoporosis, infertility, and neurological disorders such as ataxia and peripheral neuropathy. This wide spectrum of symptoms differ not only in type (digestive vs non-digestive) but also in severity, ranging from fatigue and mild discomfort to severe malabsorption syndromes; and these presentations of CD often overlaps with other gastrointestinal and systemic disorders, further complicating timely diagnosis[4,5].
The diagnosis of CD relies on a combination of clinical assessment, serological testing for antibodies (e.g., anti-tissue transglutaminase and anti-endomysial antibodies), and histological examination of duodenal biopsies, which typically reveal villous atrophy, crypt hyperplasia, and intraepithelial lymphocytosis[6,7]. Advances in non-invasive diagnostic tools, including genetic testing for human leucocyte antigen (HLA) alleles HLA-DQ2 and HLA-DQ8, have further improved the accuracy of CD diagnosis. A meta-analysis with systematic review was published including 24 studies on the association between HLA-DQB102 gene doses and the clinical and histological characteristics of CD (including clinical presentation, histology, age at diagnosis, and comorbidities). Classical CD (digestive form) was more prevalent in patients with a double dose of HLA-DQB102 vs a single allele (Odds ratio: 1.758)[8].
The prevalence of CD shows significant geographic variability. For instance, the Saharawi refugee population in Algeria has the highest recorded prevalence, affecting nearly 6% of the population[9]. In contrast, in Asia, the sero
These differences may be attributed to genetic, environmental, and dietary factors, as well as variations in diagnostic practices and awareness levels.
The recent advent of artificial intelligence (AI) is revolutionizing CD management by enabling more accurate diagnosis, risk prediction, and personalized treatment strategies. These advanced technologies represent a paradigm shift in how we approach this complex autoimmune disorder[12-16]: AI-driven models, leveraging machine learning (ML) and deep learning (DL), can analyze large-scale genomic, serological, and clinical data to improve diagnostic accuracy and identify at-risk individuals even before symptom onset.
AI on precision medicine, whereby to facilitate the gene and environment determinants of gluten intolerance identification during digestive diagnosis and predictive algorithms for improved disease subtype (i.e. genotypes) classifications or treatments outcomes forecasts to personalize recommendations[17].
Precision medicine refers to a broad approach for tailoring healthcare based on individual variability in genes, environment, and lifestyle factors. Recent technological developments have greatly improved the precision medicine approaches for CD: Next-generation sequencing is establishing comprehensive genomic profiles; proteomic technologies enables high-throughput biomarker discovery; and multiplex serological assays eases simultaneous assessment of various autoantibodies providing a more nuanced understanding of the immune responses in CD[18-20].
Actually, precision medicine would optimize the diagnosis, treatment, and prevention of diseases like CD[14-16] through three main goals:
Identifying individuals at high risk of developing CD through genetic and serological screening.
Predicting disease progression and potential complications using advanced modeling techniques.
Developing tailored management plans based on individual clinical profiles and genetic information.
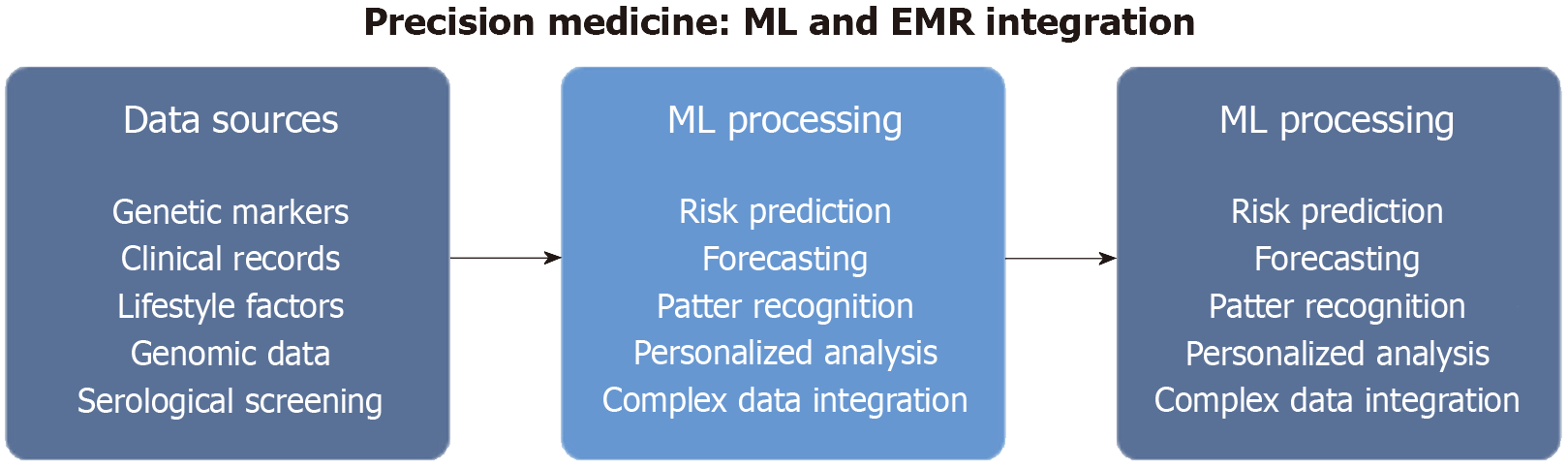
Recent advances in technology, including the use of ML and electronic medical records (EMRs), would facilitate the application of precision medicine in CD by integrating diverse data types such as genetic markers, clinical records, and lifestyle factors[21]. The Figure 1 summarizes the ML and EMR integration in Precision Medicine.
Beyond these topics related to precision medicine and ML, AI has also been used in CD to utilize different data modalities, including genomic data, EMR, and textual data, to provide personalized therapy[15].
EMRs are digital systems that store patient health information, including medical history, laboratory results, imaging studies, and treatment outcomes[22].
Integrating AI with EMRs can deliver real-time, data-driven recommendations and provide a rich resource for advancing precision medicine in CD by enabling data integration, risk stratification and patients monitoring:
Combining genetic data, biomarkers, and clinical information to create a comprehensive patient profile. This integration supports more accurate diagnosis and personalized management[23].
Identifying individuals at high risk for CD through analysis of EMR data[24], including abnormal laboratory results (i.e. genetics or autoimmunity) or patient’s patterns (age, sex, clinical symptoms, associated autoimmunity etc.).
Tracking disease progression and response to therapy over time (e.g., gluten-free diet, micro- and macro-nutrient assessments, and supplementation) and guiding adjustments in management plans[25].
Indeed, a pioneer study from the Mayo clinic (Rochester, Minnesota, United States) showed that computerized EMR-based algorithms can help identify patients at high risk of CD, and identified individuals at high risk of CD and those who would benefit from CD screening[26].
Finally, ML models can process vast amounts of EMR data to predict disease onset, detect early symptoms, and recommend targeted interventions. This computational approach transforms routine clinical data into actionable insights that support precision medicine initiatives. The integration of EMR systems with ML algorithms creates a powerful framework for precision medicine in CD. EMRs serve as the foundation by providing structured clinical data, while ML techniques analyze these datasets to identify patterns and make predictions. For example, natural language processing applications can extract relevant information from clinical notes, radiology reports, and pathology findings to create comprehensive patient profiles that inform personalized treatment decisions[27].
ML is a subset of AI that uses algorithms to analyze complex datasets and make predictions or decisions without explicit programming[15,26].
In the specific context of CD, evidence has proven that ML is efficient in several ways:
ML models can analyze clinical, genetic, and environmental data to predict the likelihood of developing CD, facilitating earlier diagnosis and intervention[28]. Recent studies demonstrate that ML models achieve up to 90% accuracy in predicting CD development in high-risk populations, compared to 64% accuracy with traditional risk assessment methods[29]. A recent convolutional neural network applied to biopsy images showed an accuracy of predicting CD at 99.97%[30].
ML algorithms can identify genetic markers associated with CD, such as HLA-DQ2 and HLA-DQ8, and predict an individual’s genetic susceptibility[31].
CD presents in various forms, including classical, atypical, and potential CD. ML techniques can analyze patient data to differentiate subtypes, enabling tailored treatment plans[15].
By analyzing longitudinal datasets, ML models can predict disease progression, guiding clinicians in monitoring and managing patients effectively[32].
For CD patients, adhering to a gluten-free diet is critical, and ML-powered tools can easily identify hidden gluten in food products, recommend safe alternatives, and provide personalized dietary advice[33]. Advanced ML techniques such as computer vision and natural language processing are being deployed in smartphone applications that can analyze food labels or restaurant menu descriptions to detect potential gluten content. These applications use deep learning models trained on extensive food databases to identify ingredients that might contain hidden gluten with over 90% accuracy, providing real-time guidance for patients managing strict gluten-free diets[34,35].
ML can stratify patients based on disease severity, response to treatment, and risk factors, optimizing resource allocation and therapeutic strategies[36].
ML can analyze big datasets to identify new drug targets, predict responses to existing treatments, and accelerate the development of novel therapies for CD[37].
ML-driven applications, such as chatbots or mobile apps, offer CD patients personalized information, emotional support, and dietary tips, improving their quality of life[38] (Table 1).
| Machine learning application in celiac disease | Description |
| Screening and early diagnosis | Predicting CD likelihood using clinical and genetic data, enabling earlier intervention |
| Genetic analysis | Algorithms identify genetic markers (HLA-DQ2/DQ8) to assess susceptibility to CD |
| Subtype identification | Differentiating between CD subtypes for personalized treatment plans |
| Disease progression prediction | Analyzing datasets helps predict disease progression and guide patient management |
| Dietary management | Identifying gluten in foods and providing personalized dietary recommendations |
| Patient stratification | Stratifying patients by severity and treatment response, optimizing care strategies |
| Drug discovery | Analyzing data to identify new drug targets and predict treatment responses for CD therapies |
| Patient support | Personalized information and support, enhancing patient quality of life |
The success of ML models depends on the availability of high-quality, diverse datasets that reduce biases and ensure accuracy. Effective clinical integration of ML also requires rigorous validation, ongoing monitoring, and robust oversight to ensure models deliver safe, reliable, and equitable care[12,38].
The application of ML in CD leverages various algorithmic approaches tailored to specific clinical questions:
Supervised learning methods, particularly support vector machines and random forests, have demonstrated particular utility in classifying CD subtypes based on clinical features. Meanwhile, deep learning neural networks excel at analyzing complex histopathological images to detect subtle tissue changes indicative of CD[30,32].
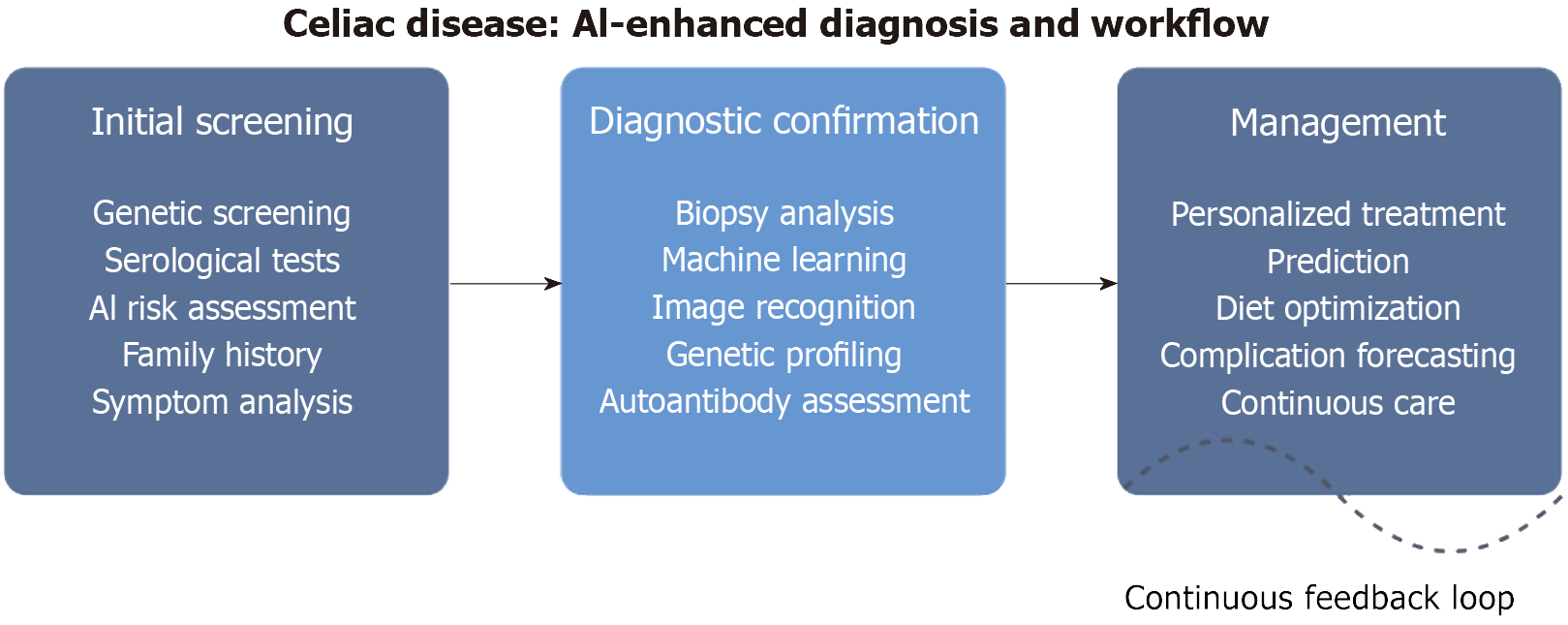
Unsupervised learning techniques, including clustering algorithms, have successfully identified novel patient subgroups with distinct clinical trajectories and treatment responses[23]. The Figure 2 depicts the AI-enhanced diagnosis and workflow in celiac disease.
Precision medicine does hold great promise for advancing the diagnosis and management of CD, but several challenges are still remaining[29,30]:
To ensure the accuracy, consistency, and completeness of datasets used for ML training and EMR analyses is mere critical.
Ethical considerations extend beyond basic privacy concerns into complex issues of informed consent, algorithmic transparency, and equitable access. Healthcare AI systems must comply with regulations such as the General Data Protection Regulation in Europe and the Health Insurance Portability and Accountability Act in the United States. Furthermore, ensuring that patients fully understand how their data will be used in ML models presents unique challenges in the digital health era[39,40].
The potential for algorithmic bias—where ML models may perform differently across demographic groups—requires rigorous validation across diverse populations to ensure that precision medicine approaches benefit all CD patients equitably[41].
The successful integration of such advanced technologies into routine clinical practice requires a continuous training of healthcare practitioners and a robust validation of these tools in diverse ethnies and populations[42]. Addressing these challenges would pave the way for broader implementation of precision medicine for celiac patients.
Precision medicine is revolutionizing CD management through the integration of EMRs and ML technologies, enabling earlier detection, personalized therapies, and enhanced disease monitoring. These advanced computational approaches leverage diverse data sources to optimize diagnostic accuracy, treatment selection, and outcome prediction. Future research should focus on external validation of ML models across diverse populations, implementation strategies for clinical integration, and development of accessible tools that bridge the technological divide. By addressing these challenges, we can realize the full potential of precision medicine to transform CD care and ultimately improve patient outcomes worldwide.
H. Rahmoune and N. Boutrid, from LIRSSEI research laboratory, are supported by the Directorate-General for Scientific Research and Technological Development (DGRSDT), MESRS, Algeria. The sponsor had no involvement in the collection, analysis and interpretation of data or the writing of the manuscript.
| 1. | Husby S, Koletzko S, Korponay-Szabó I, Kurppa K, Mearin ML, Ribes-Koninckx C, Shamir R, Troncone R, Auricchio R, Castillejo G, Christensen R, Dolinsek J, Gillett P, Hróbjartsson A, Koltai T, Maki M, Nielsen SM, Popp A, Størdal K, Werkstetter K, Wessels M. European Society Paediatric Gastroenterology, Hepatology and Nutrition Guidelines for Diagnosing Coeliac Disease 2020. J Pediatr Gastroenterol Nutr. 2020;70:141-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 794] [Article Influence: 132.3] [Reference Citation Analysis (0)] |
| 2. | Catassi C, Gatti S, Fasano A. The new epidemiology of celiac disease. J Pediatr Gastroenterol Nutr. 2014;59 Suppl 1:S7-S9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 3. | Lebwohl B, Sanders DS, Green PHR. Coeliac disease. Lancet. 2018;391:70-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 696] [Article Influence: 87.0] [Reference Citation Analysis (1)] |
| 4. | Rubio-Tapia A, Hill ID, Kelly CP, Calderwood AH, Murray JA; American College of Gastroenterology. ACG clinical guidelines: diagnosis and management of celiac disease. Am J Gastroenterol. 2013;108:656-76; quiz 677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1173] [Cited by in RCA: 1183] [Article Influence: 91.0] [Reference Citation Analysis (1)] |
| 5. | Boutrid N, Amrane M, Bioud B, Rahmoune H. Diagnosis of Celiac Disease: New Paradigms. Batna J Med Sci. 2021;8:145-150. [RCA] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Hill ID, Dirks MH, Liptak GS, Colletti RB, Fasano A, Guandalini S, Hoffenberg EJ, Horvath K, Murray JA, Pivor M, Seidman EG; North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. Guideline for the diagnosis and treatment of celiac disease in children: recommendations of the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr. 2005;40:1-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 697] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 7. | Ludvigsson JF, Bai JC, Biagi F, Card TR, Ciacci C, Ciclitira PJ, Green PH, Hadjivassiliou M, Holdoway A, van Heel DA, Kaukinen K, Leffler DA, Leonard JN, Lundin KE, McGough N, Davidson M, Murray JA, Swift GL, Walker MM, Zingone F, Sanders DS; BSG Coeliac Disease Guidelines Development Group; British Society of Gastroenterology. Diagnosis and management of adult coeliac disease: guidelines from the British Society of Gastroenterology. Gut. 2014;63:1210-1228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 689] [Cited by in RCA: 809] [Article Influence: 67.4] [Reference Citation Analysis (3)] |
| 8. | Bajor J, Szakács Z, Farkas N, Hegyi P, Illés A, Solymár M, Pétervári E, Balaskó M, Pár G, Sarlós P, Szűcs Á, Czimmer J, Szemes K, Huszár O, Varjú P, Vincze Á. Classical celiac disease is more frequent with a double dose of HLA-DQB1*02: A systematic review with meta-analysis. PLoS One. 2019;14:e0212329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Catassi C, Rätsch IM, Gandolfi L, Pratesi R, Fabiani E, El Asmar R, Frijia M, Bearzi I, Vizzoni L. Why is coeliac disease endemic in the people of the Sahara? Lancet. 1999;354:647-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 174] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 10. | Kang JY, Kang AH, Green A, Gwee KA, Ho KY. Systematic review: worldwide variation in the frequency of coeliac disease and changes over time. Aliment Pharmacol Ther. 2013;38:226-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 179] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 11. | Cabrera-Chávez F, Granda-Restrepo DM, Arámburo-Gálvez JG, Franco-Aguilar A, Magaña-Ordorica D, Vergara-Jiménez Mde J, Ontiveros N. Self-Reported Prevalence of Gluten-Related Disorders and Adherence to Gluten-Free Diet in Colombian Adult Population. Gastroenterol Res Pract. 2016;2016:4704309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 12. | Sana MK, Hussain ZM, Shah PA, Maqsood MH. Artificial intelligence in celiac disease. Comput Biol Med. 2020;125:103996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Tenório JM, Hummel AD, Cohrs FM, Sdepanian VL, Pisa IT, de Fátima Marin H. Artificial intelligence techniques applied to the development of a decision-support system for diagnosing celiac disease. Int J Med Inform. 2011;80:793-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 14. | Stoleru CA, Dulf EH, Ciobanu L. Automated detection of celiac disease using Machine Learning Algorithms. Sci Rep. 2022;12:4071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (1)] |
| 15. | Piccialli F, Calabrò F, Crisci D, Cuomo S, Prezioso E, Mandile R, Troncone R, Greco L, Auricchio R. Precision medicine and machine learning towards the prediction of the outcome of potential celiac disease. Sci Rep. 2021;11:5683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 16. | Carreras J. Artificial Intelligence Analysis of Celiac Disease Using an Autoimmune Discovery Transcriptomic Panel Highlighted Pathogenic Genes including BTLA. Healthcare (Basel). 2022;10:1550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Johnson KB, Wei WQ, Weeraratne D, Frisse ME, Misulis K, Rhee K, Zhao J, Snowdon JL. Precision Medicine, AI, and the Future of Personalized Health Care. Clin Transl Sci. 2021;14:86-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 684] [Article Influence: 114.0] [Reference Citation Analysis (0)] |
| 18. | Shoaran M, Sabaie H, Mostafavi M, Rezazadeh M. A comprehensive review of the applications of RNA sequencing in celiac disease research. Gene. 2024;927:148681. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 19. | Palanski BA, Weng N, Zhang L, Hilmer AJ, Fall LA, Swaminathan K, Jabri B, Sousa C, Fernandez-Becker NQ, Khosla C, Elias JE. An efficient urine peptidomics workflow identifies chemically defined dietary gluten peptides from patients with celiac disease. Nat Commun. 2022;13:888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 20. | Mentzer AJ, Brenner N, Allen N, Littlejohns TJ, Chong AY, Cortes A, Almond R, Hill M, Sheard S, McVean G; UKB Infection Advisory Board, Collins R, Hill AVS, Waterboer T. Identification of host-pathogen-disease relationships using a scalable multiplex serology platform in UK Biobank. Nat Commun. 2022;13:1818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 21. | Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med. 2022;28:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 1174] [Article Influence: 293.5] [Reference Citation Analysis (0)] |
| 23. | Hartmann Tolić I, Habijan M, Galić I, Nyarko EK. Advancements in Computer-Aided Diagnosis of Celiac Disease: A Systematic Review. Biomimetics (Basel). 2024;9:493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 24. | Shemesh O, Polak P, Lundin KEA, Sollid LM, Yaari G. Machine Learning Analysis of Naïve B-Cell Receptor Repertoires Stratifies Celiac Disease Patients and Controls. Front Immunol. 2021;12:627813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 25. | Månsson AL, Meijer-Boekel C, Mårild K. Utilization and Effectiveness of eHealth Technology in the Follow-up of Celiac Disease: A Systematic Review. J Pediatr Gastroenterol Nutr. 2022;74:812-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 26. | Ludvigsson JF, Pathak J, Murphy S, Durski M, Kirsch PS, Chute CG, Ryu E, Murray JA. Use of computerized algorithm to identify individuals in need of testing for celiac disease. J Am Med Inform Assoc. 2013;20:e306-e310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Latif J, Xiao C, Tu S, Rehman SU, Imran A, Bilal A. Implementation and Use of Disease Diagnosis Systems for Electronic Medical Records Based on Machine Learning: A Complete Review. IEEE Access. 2020;8:150489-150513. [DOI] [Full Text] |
| 28. | Syed S, Al-Boni M, Khan MN, Sadiq K, Iqbal NT, Moskaluk CA, Kelly P, Amadi B, Ali SA, Moore SR, Brown DE. Assessment of Machine Learning Detection of Environmental Enteropathy and Celiac Disease in Children. JAMA Netw Open. 2019;2:e195822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 29. | Wichka I, Lai PK. Rapid discovery of Transglutaminase 2 inhibitors for celiac disease with boosting ensemble machine learning. Comput Struct Biotechnol J. 2024;23:3669-3679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 30. | Carreras J. Celiac Disease Deep Learning Image Classification Using Convolutional Neural Networks. J Imaging. 2024;10:200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (1)] |
| 31. | Abraham G, Tye-Din JA, Bhalala OG, Kowalczyk A, Zobel J, Inouye M. Accurate and robust genomic prediction of celiac disease using statistical learning. PLoS Genet. 2014;10:e1004137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 32. | Mehandiratta A, Vij N, Khanna A, Gupta P, Gupta D, Gupta AK. Prediction of Celiac Disease Using Machine-Learning Techniques. Advances in Intelligent Systems and Computing. Springer Singapore, 2020. [DOI] [Full Text] |
| 33. | Pérez-Pérez M, Ferreira T, Igrejas G, Fdez-Riverola F. A novel gluten knowledge base of potential biomedical and health-related interactions extracted from the literature: Using machine learning and graph analysis methodologies to reconstruct the bibliome. J Biomed Inform. 2023;143:104398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 34. | Shaukat H, Sultan A, Salahuddin H. Enhancing food safety: A machine learning approach for accurate detection and classification of food allergens. J Comput Biomed Inform. 2024;. |
| 35. | Elyashar A, Paradise Vit A, Sebbag G, Khaytin A, Zakai A. Automated Gluten Detection in Bread Images Using Convolutional Neural Networks. Appl Sci. 2025;15:1737. [DOI] [Full Text] |
| 36. | Gruver AM, Lu H, Zhao X, Fulford AD, Soper MD, Ballard D, Hanson JC, Schade AE, Hsi ED, Gottlieb K, Credille KM. Pathologist-trained machine learning classifiers developed to quantitate celiac disease features differentiate endoscopic biopsies according to modified marsh score and dietary intervention response. Diagn Pathol. 2023;18:122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 37. | Shen T, Wang H, Hu R, Lv Y. Developing neural network diagnostic models and potential drugs based on novel identified immune-related biomarkers for celiac disease. Hum Genomics. 2023;17:76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Sadeghi P, Karimi H, Lavafian A, Rashedi R, Samieefar N, Shafiekhani S, Rezaei N. Machine learning and artificial intelligence within pediatric autoimmune diseases: applications, challenges, future perspective. Expert Rev Clin Immunol. 2024;20:1219-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 39. | Forcier MB, Gallois H, Mullan S, Joly Y. Integrating artificial intelligence into health care through data access: can the GDPR act as a beacon for policymakers? J Law Biosci. 2019;6:317-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 40. | Humphrey BA. Data privacy vs. innovation: A quantitative analysis of artificial intelligence in healthcare and its impact on HIPAA regarding the privacy and security of protected health information. MD Thesis, Robert Morris University, 2021. Available from: https://search.proquest.com/openview/146e12c7a1ead3fc5e2e4d84f67c02fd/1?pq-origsite=gscholar&cbl=18750&diss=y. |
| 41. | Akter S, Dwivedi YK, Sajib S, Biswas K, Bandara RJ, Michael K. Algorithmic bias in machine learning-based marketing models. J Bus Resh. 2022;144:201-216. [DOI] [Full Text] |
| 42. | Khoury MJ, Bowen S, Dotson WD, Drzymalla E, Green RF, Goldstein R, Kolor K, Liburd LC, Sperling LS, Bunnell R. Health equity in the implementation of genomics and precision medicine: A public health imperative. Genet Med. 2022;24:1630-1639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/