Published online Oct 28, 2021. doi: 10.35713/aic.v2.i5.69
Peer-review started: October 19, 2021
First decision: October 24, 2021
Revised: October 24, 2021
Accepted: October 27, 2021
Article in press: October 27, 2021
Published online: October 28, 2021
Processing time: 8 Days and 22.4 Hours
With the rapid development of high-throughput sequencing and artificial intelligence (AI) techniques, gut mucosal microbiota begins to be recognized as critical drivers of human colorectal cancer (CRC). Various AI approaches have been designed to obtain effective information from enormous numbers of microbial cells residing in gut mucosal as well as cancer cells. These mainly include detection of microbial markers for early clinical diagnosis of stage-specific CRC, characterization of pathogenic bacterial activities via genomic and transcriptomic analyses, and prediction of interplay between bacterial drivers and host immune systems. Here I review the current progresses of AI applications in profiling gut microbiomes linked to CRC initiation and development. I further look forward to future AI research for improving our understanding of the roles of gut microbiota in CRC evolution.
Core Tip: In this review, the author reviews the current progresses of artificial intelligence (AI) applications in profiling gut microbiomes linked to colorectal cancer (CRC) initiation and development. The author further looks forward to future AI research for improving our understanding of the roles of gut microbiota in CRC evolution.
- Citation: Wan XH. Artificial intelligence reveals roles of gut microbiota in driving human colorectal cancer evolution. Artif Intell Cancer 2021; 2(5): 69-78
- URL: https://www.wjgnet.com/2644-3228/full/v2/i5/69.htm
- DOI: https://dx.doi.org/10.35713/aic.v2.i5.69
Colorectal cancer (CRC) continuously receives public and academic attentions due to its high prevalence and mortality rate[1]. Understanding the genetic mechanisms behind CRC initiation and progression is important to the development of early diagnosis and new therapy for CRC and its recurrence. The concept of the adenoma-carcinoma sequence, which refers to a sequential activation of oncogenes and inactivation of tumor suppressor genes, is well recognized for CRC progression[2,3]. The adenoma-carcinoma sequence involves genetic mutations and epigenetic modification of human genome in vivo, which have been believed to be caused by exogenous and endogenous mutagens for decades[4-6]. However, it is still not fully understood which exogenous mutagens induce cancers and the induction mechanisms behind them remain largely unknown, especially when the questions go deep to a defined type of cancer.
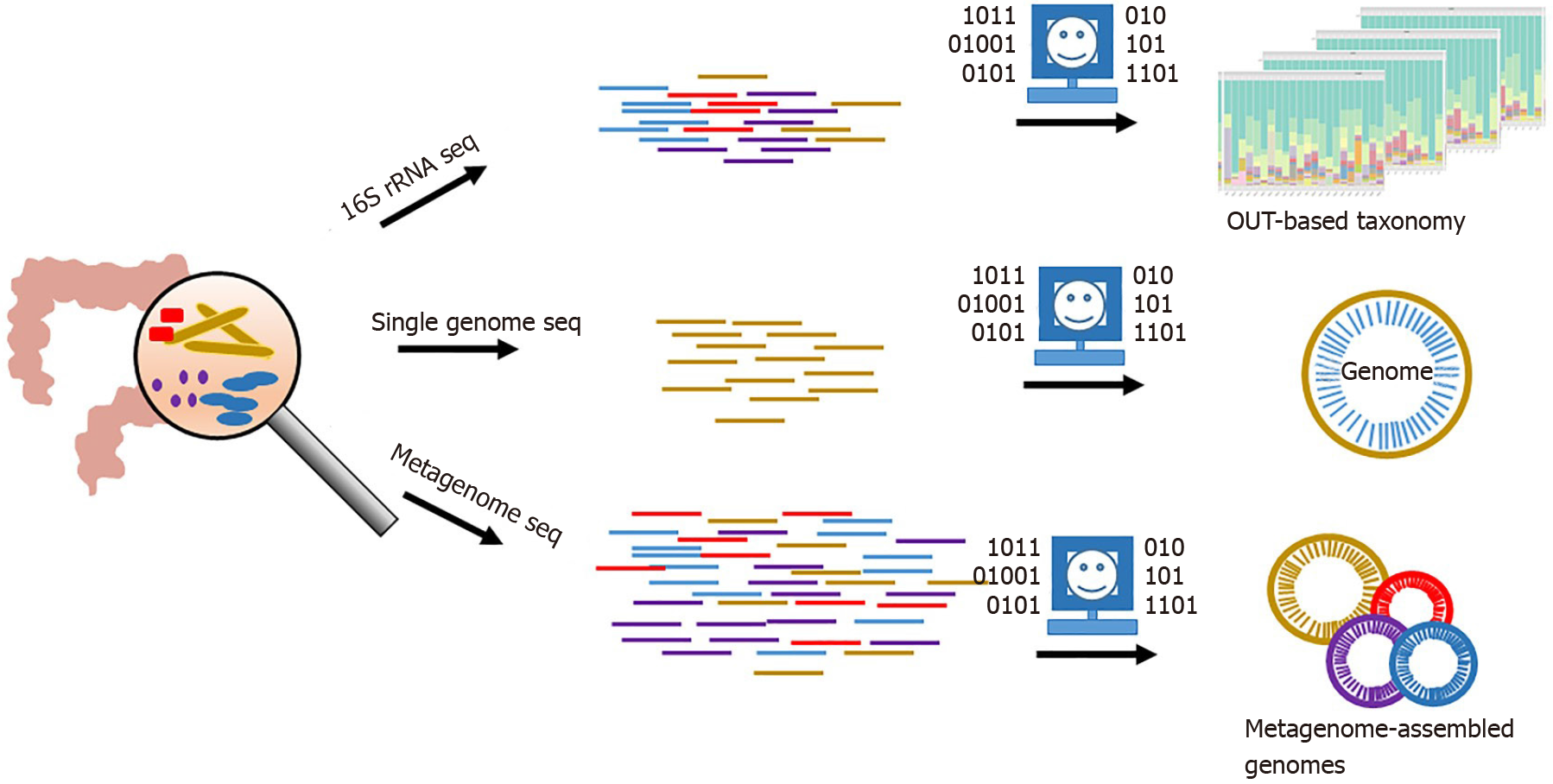
Growing evidences indicate that gut mucosal microbiota is strongly linked to CRC development and may serve as a primary driver to induce inflammation in the human colon[7-13]. High-throughput sequencing (HTS) of 16S ribosomal RNA (rRNA) gene fragments is widely applied to profile microbial communities and used to study the composition structures of gut mucosal microbiota associated with human CRC (Figure 1)[14-17]. Moreover, metagenome sequencing of gut mucosal microbiomes coupled with binning strategies and other downstream analysis are able to reveal metabolism pathways in potential pathogenic bacteria at lineage levels, which are critical to screening microbial biomarkers (e.g., taxa and gene) for CRC and understanding the microbe-host interactions (Figure 1)[18-20]. Emerging meta transcriptomic sequencing, which examines large-scale gene expressions in microbial communities, is able to provide comprehensive insights into microbial population activities in host. Based on these in silico analyses and following wet-lab validations, species such as Fusobacterium nucleatum, Peptostreptococcus anaerobius, pks+ Escherichia coli and Eubacterium rectale have been identified as pathogenic drivers responsible for CRC progression[9,10,12,21]. However, due to the expensive and time-consuming wet-lab experiments, a list of CRC-associated species is on the way to be examined for the physiological roles in CRC progression. Instead, AI approaches can serve as efficient methods to detect potential roles of these microbes in microbe-host interactions and provide clues for wet-lab validation.
With its increasingly wide applications in our everyday life, e.g. self-driving cars, facial recognition, and medical diagnosis, AI becomes one of the most popular fields that are heavily invested and supported in a number of countries. AI is capable of mimicking and going beyond human capabilities. In some biological fields such as genomics and transcriptomics, AI is able to complete the complex tasks that are impossible for human to finish[22]. AI technique encompasses machine learning (ML) as a major branch that includes deep learning as a subset of ML[23,24]. In essence, ML are computing algorithms that are either supervised by training datasets or designed as unsupervised algorithms. They are widely applied in gut microbiome field. Here I review the current progresses of AI applications in detection of pathogenic drivers for CRC and prediction of their driving roles in CRC evolution.
To understand the roles of pathogenic bacterial species in initiating and driving CRC progression, the first and most important step is to identify the spectrum of indigenous bacterial taxonomy in human gut. Current HTS technology has developed sufficiently mature methods and is able to extensively characterize bacterial taxonomy in samples collected from diverse environments and various hosts, including human gut mucosal[14-20,25,26]. As a key step for taxonomic assignment, classification of operational taxonomic units (OTUs) from large datasets of HTS 16S rRNA sequencing reads employs various AI algorithms. Classical algorithms for OTU classification include long-sequence-fist list removal algorithm[27,28], uclust algorithm[29], random forest algorithm[30], and RDP naïve Bayesian classifier algorithm[31]. Because the datasets are usually generated in large scales, both accuracy and computation speed must be considered for trade off. Long-sequence-fist list removal algorithm implements a super-fast heuristic to identity DNA segments with high identity between sequences, to avoid costly computational alignments of full sequences[27,28]. Uclust algorithm sorts k-mer of sequencing reads to rapidly identify sequences in common[29]. Random forest algorithm builds an ensemble of decision trees that are trained with a combination of learning models[30]. RDP naïve Bayesian classifier algorithm classifies based on the multinomial model in both training and testing for computing classification probabilities[31]. However, challenges still remain to accurately determine the species using 16S rRNA sequences. Errors introduced due to experimental limitations such as polymerase chain reaction amplification and HTS sequencing need to be considered. In addition, although hypervariable regions in 16S rRNA sequences were used for taxonomic assignment, some sequences from bacterial species within the same genus are highly homologous or identical, leading to problems for taxonomic assignment. To solve these issues, new algorithms are also developed. For example, Bayesian-like operational taxonomic unit examiner algorithm employs a grammar-based assignment strategy to deal with sequencing reads errors, in which unsupervised Bayesian models are built based on k-mers split from sequencing reads[32]. To solve homology issues of hypervariable regions in 16S rRNA, Gwak and Rho used a k-nearest neighbor algorithm and the species consensus sequence models to determine species-level taxonomy[33]. Further development of AI methods for OTU classification will help improve the accuracy for taxonomic assignment and speed for dealing with large-scale dataset.
Since gut microbiome OTUs may represent novel species/strains, placing them on a phylogenetic tree can shed light on their taxonomic positions. The computation of phylogenetic likelihood for reconstruction of evolutionary tress from sequence data is both memory and computing consuming. Both Neighbor-Joining (NJ) and maximum-likelihood algorithms are the most popular methods in resolving topology of OTU sequences[34-38]. The NJ tree inference method belongs to distance-based method and takes a matrix of pairwise distance between the sequences to build evolutionary tree. The maximum-likelihood algorithm calculates all the possible tree topologies based on the probability.
The composition structure of gut microbiome is highly complex, containing high-dimensional information for hundreds of bacterial species and their abundances[39]. To apply data mining strategies on looking for critical factors that distinguish gut microbiomes, large numbers of samples were usually collected from patients in different CRC conditions, such as various intestinal locations and CRC stages. To examine the differences among samples that belong to specific conditions, the high-dimensional information from each sample need to be reduced and presented on a two-dimensional space. As an unsupervised algorithm, principal component analysis is a dimensionality reduction algorithm that transforms and compresses matrix consisting of high-dimensional interrelated variables to a new set of two-dimensional variables[40,41]. By plotting the compressed two-dimensional variables, the microbiome patterns of gut mucosal samples collected from different conditions can be evaluated.
To understand gut microbiome functions, bacteria residing in gut mucosal ecosystem need to be isolated and cultivated in laboratory for experimental validation[42]. Sequencing the genomes of these bacteria can reveal their metabolism traits and guide downstream functional analyses. For whole genome shotgun sequencing, bacterial genomic DNA is fragmented into small pieces for 2 × 100 or 2 × 150 bp paired-end sequencing. Various de novo assemblers, including Velvet, SPAdes and SoapDeNovo, have been designed to assemble a large number of short sequence reads to form a set of contiguous sequences representing the genome[43-45]. Because the reads are short, they are usually generated in large quantities with a high coverage depth. To deal with such a large dataset, the assemblers are not designed to assemble the short reads directly. Instead, the reads are splitted to form a set of k-mers and then mapped through de Bruijn graph. Although de Bruijn graph is suggested for short read assembly (100-200 bp), it is not recommended to assemble very short reads (25-50 bp). Velvet was designed to manipulate de Bruijn graph algorithm efficiently for very short reads assembly[43]. Elimination of errors and resolving repeats regions were considered in Velvet[43]. Reconstruction of consensus sequences from k-mers based on de Bruijn algorithm may lead to fragmented assembly. To deal with the issues, paired de Bruijn graphs using read-pairs (bireads) was designed. Inspired by paired de Bruijn graphs, SPAdes uses paired assembly graph algorithm by introducing k-bimer adjustment that reveals exact distances for the adjusted k-bimers[44]. SOAPdevo2, as the version 2 of SOAPdenovo, also utilizes de Bruijn graph algorithm but is designed to reduce memory consumption in de Bruijn graph constructions[45]. The algorithm supports error correction for long k-mers to improve accuracy and sensitivity during the assembly process. Moreover, the program benefits the assembly of repeat regions with high coverage depth and regions with low coverage depth via application of a k-mer size selection strategy. Therefore, these assembly algorithms have their specific advantages and are widely utilized in practical applications.
Gut mucosal microbiomes comprise hundreds of bacterial species, of which some are uncultivable in laboratory conditions[46,47]. Sequencing these mixed bacterial populations facilitates discovery of the genomic traits of these uncultivable bacteria. Although assembling the reads and reconstructing genes from these complex mixtures are challenging, metagenomic assembly algorithms and downstream binning strategies are under developing progresses to solve the technique problems.
Genome assembly for sequencing reads from a single species assumes that all the reads are sequenced from the same genomic DNA and contaminations can be screened out during quality control process[48]. The genome size of single species can be estimated based on the sizes of close phylogenetic neighbors and k-mer counting, and the required sequencing depth can be calculated according to the genome size. During assembly process, de Bruijn algorithm is designed to simply consider nodes or edges with low coverage depth as contamination and remove them[48,49]. In the same way, nodes with high coverage depth are considered by the algorithm as repetitive regions in the genome sequence. In contrast, metagenomic assembly cannot make such a simple assumption to decide nodes with low and high coverage depths to be from contamination sequences or repetitive regions. This is because metagenomic sequencing reads are generated from mixed bacterial populations, in which certain species grow better than the rest and show high abundances in the mixed communities, whereas rare species show low abundances. Therefore, the coverage depths of heterogeneous reads cannot facilitate the assumption of their origins.
Currently, the most popular assemblers for metagenomics assembly include MEGAHIT and metaSPAdes[50,51]. MEGAHIT utilizes a fast parallel algorithm for succinct de Bruijn graphs to assemble k-mers from metagenomics reads[50]. To avoid k-mer singletons caused by sequencing error, MEGAHIT sorts and counts all (k + 1)-mers splitted from the sequencing reads and only counts (k + 1)-mers with > 2 occurrences[50]. In addition, MEGAHIT utilizes a mercy-kmers strategy to recover low-depth edges for the assembly of rare species[50]. MetaSPAdes uses de Bruijn graph of all reads using SPAdes, transforms it into the assembly graph using various simplification procedures[51]. The algorithm works across a wide range of coverage depths.
Since assembled metagenomic scaffolds/contigs are derived from each species and show sequence composition characteristics such as GC content and coverage depth, various binning strategies are designed for the reconstruction of metagenome-assembled genome (MAG). MAGs represent genomes from monophyletic lineages and can be used to analyze taxonomic and metabolic potentials. A number of programs have been designed for MAG binning, including MetaBat2, Maxbin2, CONCOCT, MyCC, and BinSanity[52-56]. MetaBat2 is a user-friendly program that does not need to tune the parameters for its sensitivity and specificity[52]. It utilizes a new adaptive binning algorithm to tune these parameters automatically, and uses a graph based structure for contig clustering. MetaBat2 is optimized for extensive low-level computation and works very efficiently for very large datasets. MaxBin 2.0 employs an Expectation-Maximization algorithm to recover draft genomes from metagenomes[53]. It measures the tetranucleotide frequencies of the contigs and their coverages and then classifies the contigs into each bins. CONCOCT uses Gaussian mixture models to cluster contigs into bins[54]. Sequence composition and coverage are considered for assigning contigs to bins. A variational Bayesian approach is used to determine the number of clusters. MyCC works in a way using metagenomics signatures, contig/scaffold coverage depths, and Barnes-Hut-SNE-based dimension reduction[55]. MyCC predicts genes in metagenomic contigs using Prodigal and then identifies single-copy marker genes using Hidden Markov Model trained FetchMG along with UCLUST. The reduced genomic signatures via Barnes-Hut-SNE algorithm are then clustered using affinity propagation for binning. Similarly, BinSanity utilizes affinity propagation algorithm to generate bins based on coverage depth, tetranucleotide frequency, and GC content[56]. Although these bin extraction algorithms are designed based on their own specific principles, the resulted bins from the same dataset can be combined, evaluated, modified, and improved to generate high-quality final set of bins using metaWRAP[57].
Quality evaluation of the assembled MAGs determines the reliability of downstream annotation analyses. Because the concept of metagenome sequencing is quite new, not many programs have been developed with matured principles to determine MAG qualities. Currently, the most popular program is CheckM, which uses a set of lineage-specific marker genes within a reference genome tree[58]. By this way, CheckM estimates the completeness and contamination of the assembled MAGs and determines which MAGs are useful for downstream analyses. To determine the set of marker genes, CheckM reconstructed a genome tree based on 5656 reference genomes and then inferred the marker gene set using HMMER based on hidden Markov models and FastTree based on WAG and GAMMA models. To evaluate a MAG, the marker gene set is identified in the MAG using hidden Markov models. The identified homologous genes of the marker genes are further aligned, concatenated, and then placed into the reference genome tree using pplacer for taxonomic inference and quality checking[59]. Another evaluation method for the assembled MAG is MetaQUAST, which aligns contig sequences of MAG to a close reference genome[60]. This program is able to detect potential taxonomic position of MAG by BLASTN searches against 16S rRNA sequences from the SILVA database[61,62]. Then it automatically downloads close reference genomes from the on-line NCBI database and aligns them against MAG for evaluation.
Different from the taxonomic assignment based on 16S rRNA sequencing, metagenome sequencing and assembly contain much more information than 16S rRNA sequences. Data mining strategies to obtain taxonomic information from large-scale metagenome assembly need to be considered and designed. As discussed above, both CheckM and MetaQUAST provide lineage hints for taxonomic assignment of MAGs[58,60]. Additionally, PhyloFlash maps sequencing reads to small-subunit rRNA (SSU rRNA) database for taxonomic assignment and can be performed before the metagenomes are assembled[63]. FOCUS uses non-negative least squares algorithm to compare k-mers between references genomes and MAGs, and determine taxonomic position for contigs binned in MAGs[64].
Gut microbes living in intestine mucosal, including commensals and pathogens, regulate homeostasis of host immunity[65]. Their activities are able to alter host signaling and immunity by interacting with the host proteins. Deciphering how microbe and host interact via protein-protein interactions and through which microbial and host proteins they work are important to development of novel strategies for prevention of CRC. Since wet-lab experiments are time-consuming and laborious, experimentally determining the microbe-host interactions is still challenging. On the other hand, genome-wide computational methods can efficiently provide hints to enhance our understanding of this challenging task[66-71]. One category of these computational methods are AI based methods for determining protein-protein interactions (PPI) between microbes and host[69,70]. Currently, AI based methods for PPI predictions are still new and only a few of them have been developed. Most of them are supervised methods, which utilizes well-recognized datasets as standards to train AI models and determine parameters. These training datasets are either collected from high-throughput experiments or obtained from literatures by text mining. Supervised PPI methods utilize various AI models such as logistic regression, random forests, support vector machine, artificial neural networks, and K-nearest neighbors[72-76]. However, these AI-based PPI methods are designed for the PPI relationship between specific pathogen and human such as human-Bacillus anthracis, human-Yersinia pestis and human-Fusobacterium nucleatum[67,77-79]. Because high abundances of F. nucleatum are associated with CRC patients and especially associated with specific CRC stages, F. nucleatum is proposed for its causal role in CRC development. Computational scanning of F. nucleatum genome and human proteins identified FusoSecretome proteins and their targets in the host network[67]. PPI-coupled network analysis identified that F. nucleatum perturbed host cellular pathways including immune and infection response, homeostasis, cytoskeleton organization, and gene expression regulation[67]. However, AI-based PPI studies for human-microbiome interactions still need more efforts due to the complex mixed-population of species within gut microbiome.
Rapid development of high-throughput sequencing and high-throughput screening experiments generate large-scale datasets and largely improve our understanding of functional roles of gut microbiomes in CRC evolution. Using AI-based analyses, potential pathogenic species from gut microbiome have been identified to play critical roles in driving CRC. However, there are still limitations in current methods and challenges remain for them to be improved. These include but not limited to the questions as follows. How to accurately identify bacterial species/strains that reside in gut mucosal? How to use metagenomics sequencing data to assemble complete or nearly complete MAGs for bacterial single species? How to build AI models to interpret human-microbiome interactions under different environmental conditions? And many more challenges remain to be solved. I believe that continuous improvement of AI technology in CRC diagnosis as well as many more diseases will facilitate answering the above questions and help develop clinical treatment and prevention of CRC in advance.
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56683] [Article Influence: 7085.4] [Reference Citation Analysis (135)] |
| 2. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8099] [Article Influence: 225.0] [Reference Citation Analysis (1)] |
| 3. | Smit WL, Spaan CN, Johannes de Boer R, Ramesh P, Martins Garcia T, Meijer BJ, Vermeulen JLM, Lezzerini M, MacInnes AW, Koster J, Medema JP, van den Brink GR, Muncan V, Heijmans J. Driver mutations of the adenoma-carcinoma sequence govern the intestinal epithelial global translational capacity. Proc Natl Acad Sci U S A. 2020;117:25560-25570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 4. | Morley AA, Turner DR. The contribution of exogenous and endogenous mutagens to in vivo mutations. Mutat Res. 1999;428:11-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719-724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2798] [Cited by in RCA: 2488] [Article Influence: 146.4] [Reference Citation Analysis (0)] |
| 6. | Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1515] [Cited by in RCA: 1516] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 7. | Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 692] [Article Influence: 49.4] [Reference Citation Analysis (1)] |
| 8. | Song M, Chan AT. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin Gastroenterol Hepatol. 2019;17:275-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 252] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 9. | Zhang S, Cai S, Ma Y. Association between Fusobacterium nucleatum and colorectal cancer: Progress and future directions. J Cancer. 2018;9:1652-1659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, Coker OO, Chan AWH, Chan FKL, Sung JJY, Yu J. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4:2319-2330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 367] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 11. | Rhee KJ, Wu S, Wu X, Huso DL, Karim B, Franco AA, Rabizadeh S, Golub JE, Mathews LE, Shin J, Sartor RB, Golenbock D, Hamad AR, Gan CM, Housseau F, Sears CL. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009;77:1708-1718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 233] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 12. | Wang Y, Wan X, Wu X, Zhang C, Liu J, Hou S. Eubacterium rectale contributes to colorectal cancer initiation via promoting colitis. Gut Pathog. 2021;13:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 13. | Wang Y, Zhang C, Hou S, Wu X, Liu J, Wan X. Analyses of Potential Driver and Passenger Bacteria in Human Colorectal Cancer. Cancer Manag Res. 2020;12:11553-11561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 14. | Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, He Y, Kang Q, Cao L, Wang K, Zhang J, Liang Q, Yu J, Sung JJ. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 558] [Cited by in RCA: 540] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 15. | Dadkhah E, Sikaroodi M, Korman L, Hardi R, Baybick J, Hanzel D, Kuehn G, Kuehn T, Gillevet PM. Gut microbiome identifies risk for colorectal polyps. BMJ Open Gastroenterol. 2019;6:e000297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 16. | Saito K, Koido S, Odamaki T, Kajihara M, Kato K, Horiuchi S, Adachi S, Arakawa H, Yoshida S, Akasu T, Ito Z, Uchiyama K, Saruta M, Xiao JZ, Sato N, Ohkusa T. Metagenomic analyses of the gut microbiota associated with colorectal adenoma. PLoS One. 2019;14:e0212406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 17. | Zhang M, Lv Y, Hou S, Liu Y, Wang Y, Wan X. Differential Mucosal Microbiome Profiles across Stages of Human Colorectal Cancer. Life (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 18. | Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, Chan FKL, Kristiansen K, Sung JJY, Wong SH, Yu J. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 250] [Cited by in RCA: 378] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 19. | Chen F, Dai X, Zhou CC, Li KX, Zhang YJ, Lou XY, Zhu YM, Sun YL, Peng BX, Cui W. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 203] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 20. | Mizutani S, Yamada T, Yachida S. Significance of the gut microbiome in multistep colorectal carcinogenesis. Cancer Sci. 2020;111:766-773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 21. | Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, Paganelli FL, Geurts MH, Beumer J, Mizutani T, Miao Y, van der Linden R, van der Elst S; Genomics England Research Consortium, Garcia KC, Top J, Willems RJL, Giannakis M, Bonnet R, Quirke P, Meyerson M, Cuppen E, van Boxtel R, Clevers H. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature. 2020;580:269-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 833] [Article Influence: 138.8] [Reference Citation Analysis (0)] |
| 22. | Sharma A, Rani R. A systematic review of applications of machine learning in cancer prediction and diagnosis. Arch Comput Methods Eng. 2021;. [DOI] [Full Text] |
| 23. | Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1362] [Cited by in RCA: 1378] [Article Influence: 114.8] [Reference Citation Analysis (0)] |
| 24. | Choi RY, Coyner AS, Kalpathy-Cramer J, Chiang MF, Campbell JP. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl Vis Sci Technol. 2020;9:14. [PubMed] |
| 25. | Pallen MJ, Loman NJ, Penn CW. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr Opin Microbiol. 2010;13:625-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 26. | Lightbody G, Haberland V, Browne F, Taggart L, Zheng H, Parkes E, Blayney JK. Review of applications of high-throughput sequencing in personalized medicine: barriers and facilitators of future progress in research and clinical application. Brief Bioinform. 2019;20:1795-1811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 27. | Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 682] [Cited by in RCA: 686] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 28. | Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658-1659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6482] [Cited by in RCA: 7827] [Article Influence: 391.4] [Reference Citation Analysis (4)] |
| 29. | Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460-2461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14305] [Cited by in RCA: 14325] [Article Influence: 895.3] [Reference Citation Analysis (0)] |
| 30. | Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537-7541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14372] [Cited by in RCA: 14201] [Article Influence: 835.4] [Reference Citation Analysis (0)] |
| 31. | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261-5267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12842] [Cited by in RCA: 13562] [Article Influence: 713.8] [Reference Citation Analysis (0)] |
| 32. | Koparde VN, Adkins RS, Fettweis JM, Serrano MG, Buck GA, Reimers MA, Sheth NU. BOTUX: bayesian-like operational taxonomic unit examiner. Int J Comput Biol Drug Des. 2014;7:130-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 33. | Gwak HJ, Rho M. Data-Driven Modeling for Species-Level Taxonomic Assignment From 16S rRNA: Application to Human Microbiomes. Front Microbiol. 2020;11:570825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 34. | Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3805] [Cited by in RCA: 14054] [Article Influence: 360.4] [Reference Citation Analysis (0)] |
| 35. | Mailund T, Brodal GS, Fagerberg R, Pedersen CN, Phillips D. Recrafting the neighbor-joining method. BMC Bioinformatics. 2006;7:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 36. | Dhar A, Minin VN. Maximum likelihood phylogenetic inference. Ency Evol Bio. 2016;2:499-506. [DOI] [Full Text] |
| 37. | Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7759] [Cited by in RCA: 10325] [Article Influence: 645.3] [Reference Citation Analysis (0)] |
| 38. | Sagulenko P, Puller V, Neher RA. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:vex042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 556] [Cited by in RCA: 852] [Article Influence: 106.5] [Reference Citation Analysis (0)] |
| 39. | Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474:1823-1836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1710] [Cited by in RCA: 2239] [Article Influence: 248.8] [Reference Citation Analysis (8)] |
| 40. | Swets DL, Weng J. Using discriminant eigenfeatures for image retrieval. IEEE Trans Pattern Anal Mach Intell. 1996;18:831-836. [DOI] [Full Text] |
| 41. | Turk M, Pentland A. Eigenfaces for recognition. J Cogn Neurosci. 1991;3:71-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9330] [Cited by in RCA: 2056] [Article Influence: 58.7] [Reference Citation Analysis (1)] |
| 42. | Forster SC, Kumar N, Anonye BO, Almeida A, Viciani E, Stares MD, Dunn M, Mkandawire TT, Zhu A, Shao Y, Pike LJ, Louie T, Browne HP, Mitchell AL, Neville BA, Finn RD, Lawley TD. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol. 2019;37:186-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 432] [Cited by in RCA: 385] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 43. | Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821-829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8234] [Cited by in RCA: 7250] [Article Influence: 402.8] [Reference Citation Analysis (9)] |
| 44. | Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14426] [Cited by in RCA: 19201] [Article Influence: 1371.5] [Reference Citation Analysis (0)] |
| 45. | Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3769] [Cited by in RCA: 3761] [Article Influence: 268.6] [Reference Citation Analysis (0)] |
| 46. | Kenny DJ, Plichta DR, Shungin D, Koppel N, Hall AB, Fu B, Vasan RS, Shaw SY, Vlamakis H, Balskus EP, Xavier RJ. Cholesterol Metabolism by Uncultured Human Gut Bacteria Influences Host Cholesterol Level. Cell Host Microbe. 2020;28:245-257.e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 213] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 47. | Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499-504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 557] [Cited by in RCA: 890] [Article Influence: 127.1] [Reference Citation Analysis (0)] |
| 48. | Ayling M, Clark MD, Leggett RM. New approaches for metagenome assembly with short reads. Brief Bioinform. 2020;21:584-594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (1)] |
| 49. | Sedlar K, Kupkova K, Provaznik I. Bioinformatics strategies for taxonomy independent binning and visualization of sequences in shotgun metagenomics. Comput Struct Biotechnol J. 2017;15:48-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 50. | Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674-1676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2609] [Cited by in RCA: 5675] [Article Influence: 515.9] [Reference Citation Analysis (0)] |
| 51. | Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824-834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1694] [Cited by in RCA: 2844] [Article Influence: 316.0] [Reference Citation Analysis (0)] |
| 52. | Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 791] [Cited by in RCA: 2318] [Article Influence: 331.1] [Reference Citation Analysis (0)] |
| 53. | Wu YW, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 885] [Cited by in RCA: 1703] [Article Influence: 154.8] [Reference Citation Analysis (0)] |
| 54. | Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1084] [Cited by in RCA: 1661] [Article Influence: 138.4] [Reference Citation Analysis (0)] |
| 55. | Lin HH, Liao YC. Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci Rep. 2016;6:24175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 157] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 56. | Graham ED, Heidelberg JF, Tully BJ. BinSanity: unsupervised clustering of environmental microbial assemblies using coverage and affinity propagation. PeerJ. 2017;5:e3035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 119] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 57. | Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1051] [Cited by in RCA: 1585] [Article Influence: 198.1] [Reference Citation Analysis (0)] |
| 58. | Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043-1055. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4404] [Cited by in RCA: 8511] [Article Influence: 773.7] [Reference Citation Analysis (0)] |
| 59. | Matsen FA, Kodner RB, Armbrust EV. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 786] [Cited by in RCA: 768] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 60. | Mikheenko A, Saveliev V, Gurevich A. MetaQUAST: evaluation of metagenome assemblies. Bioinformatics. 2016;32:1088-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 424] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 61. | Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590-D596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15104] [Cited by in RCA: 19735] [Article Influence: 1518.1] [Reference Citation Analysis (0)] |
| 62. | Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Res. 2014;42:D643-D648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1629] [Cited by in RCA: 2327] [Article Influence: 179.0] [Reference Citation Analysis (0)] |
| 63. | Gruber-Vodicka HR, Seah BKB, Pruesse E. phyloFlash: Rapid Small-Subunit rRNA Profiling and Targeted Assembly from Metagenomes. mSystems. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 64. | Silva GG, Cuevas DA, Dutilh BE, Edwards RA. FOCUS: an alignment-free model to identify organisms in metagenomes using non-negative least squares. PeerJ. 2014;2:e425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 65. | Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 604] [Cited by in RCA: 1219] [Article Influence: 152.4] [Reference Citation Analysis (0)] |
| 66. | Zuñiga C, Zaramela L, Zengler K. Elucidation of complexity and prediction of interactions in microbial communities. Microb Biotechnol. 2017;10:1500-1522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 67. | Zanzoni A, Spinelli L, Braham S, Brun C. Perturbed human sub-networks by Fusobacterium nucleatum candidate virulence proteins. Microbiome. 2017;5:89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 68. | Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha A, Greenhalgh K, Jäger C, Baginska J, Wilmes P, Fleming RM, Thiele I. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017;35:81-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 533] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 69. | Lian X, Yang S, Li H, Fu C, Zhang Z. Machine-Learning-Based Predictor of Human-Bacteria Protein-Protein Interactions by Incorporating Comprehensive Host-Network Properties. J Proteome Res. 2019;18:2195-2205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 70. | Guven-Maiorov E, Hakouz A, Valjevac S, Keskin O, Tsai CJ, Gursoy A, Nussinov R. HMI-PRED: A Web Server for Structural Prediction of Host-Microbe Interactions Based on Interface Mimicry. J Mol Biol. 2020;432:3395-3403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 71. | Jansma J, El Aidy S. Understanding the host-microbe interactions using metabolic modeling. Microbiome. 2021;9:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 72. | Prasasty VD, Hutagalung RA, Gunadi R, Sofia DY, Rosmalena R, Yazid F, Sinaga E. Prediction of human-Streptococcus pneumoniae protein-protein interactions using logistic regression. Comput Biol Chem. 2021;92:107492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 73. | Wei ZS, Yang JY, Shen HB, Yu DJ. A Cascade Random Forests Algorithm for Predicting Protein-Protein Interaction Sites. IEEE Trans Nanobioscience. 2015;14:746-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 74. | Chakraborty A, Mitra S, De D, Pal AJ, Ghaemi F, Ahmadian A, Ferrara M. Determining Protein–Protein Interaction Using Support Vector Machine: A Review. IEEE Access. 2021;9:12473-12490. [DOI] [Full Text] |
| 75. | Tsuchiya Y, Tomii K. Neural networks for protein structure and function prediction and dynamic analysis. Biophys Rev. 2020;12:569-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 76. | Suratanee A, Plaimas K. Reverse Nearest Neighbor Search on a Protein-Protein Interaction Network to Infer Protein-Disease Associations. Bioinform Biol Insights. 2017;11:1177932217720405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 77. | Ahmed I, Witbooi P, Christoffels A. Prediction of human-Bacillus anthracis protein-protein interactions using multi-layer neural network. Bioinformatics. 2018;34:4159-4164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 78. | Dyer MD, Neff C, Dufford M, Rivera CG, Shattuck D, Bassaganya-Riera J, Murali TM, Sobral BW. The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS One. 2010;5:e12089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 113] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 79. | Yang H, Ke Y, Wang J, Tan Y, Myeni SK, Li D, Shi Q, Yan Y, Chen H, Guo Z, Yuan Y, Yang X, Yang R, Du Z. Insight into bacterial virulence mechanisms against host immune response via the Yersinia pestis-human protein-protein interaction network. Infect Immun. 2011;79:4413-4424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Microbiology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Herold Z S-Editor: Wang JL L-Editor: A P-Editor: Wang JL