Published online Sep 18, 2023. doi: 10.13105/wjma.v11.i6.266
Peer-review started: April 4, 2023
First decision: May 15, 2023
Revised: July 15, 2023
Accepted: July 25, 2023
Article in press: July 25, 2023
Published online: September 18, 2023
Processing time: 162 Days and 0.3 Hours
In December, 2019, pneumonia triggered by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) surfaced in Wuhan, China. An acute respiratory illness named coronavirus disease 2019 (COVID-19) is caused by a new coronavirus designated as SARS-CoV-2. COVID-19 has surfaced as a major pandemic in the 21st century as yet. The entire world has been affected by this virus. World Health Organization proclaimed COVID-19 pandemic as a public health emergency of international concern on January 30, 2020. SARS-CoV-2 shares the same genome as coronavirus seen in bats. Therefore, bats might be its natural host of this virus. It primarily disseminates by means of the respiratory passage. Evidence revealed human-to-human transmission. Fever, cough, tiredness, and gastrointestinal illness are the manifestations in COVID-19-infected persons. Senior citizens are more vulnerable to infections which can lead to dangerous consequences. Various treatment strategies including antiviral thera
Core Tip: An acute respiratory illness (COVID-19) is caused by a new coronavirus designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 belongs to β-coronaviruses, and it shares the same genome as coronavirus seen in bats. It primarily disseminates by means of the respiratory passage. Much evidence revealed human-to-human transmission. Fever, cough, tiredness, and gastrointestinal illness are the manifestations in COVID-19-infected persons. Various antiviral therapies are accessible for the handling of COVID-19 disease.
- Citation: Mokhria RK, Bhardwaj JK, Sanghi AK. History, origin, transmission, genome structure, replication, epidemiology, pathogenesis, clinical features, diagnosis, and treatment of COVID-19: A review. World J Meta-Anal 2023; 11(6): 266-276
- URL: https://www.wjgnet.com/2308-3840/full/v11/i6/266.htm
- DOI: https://dx.doi.org/10.13105/wjma.v11.i6.266
In December 2019, in Wuhan (China) an outbreak of pneumonia symptomatized by fever, dry cough, fatigue, and occasional gastrointestinal symptoms was revealed. Most of these pneumonia patients were associated with the Huanan Seafood Market, Wuhan, China which deals in fish and various live animal species (poultry, bats, marmots, and snakes)[1].
By using reverse transcription polymerase chain reaction (RT-PCR), researchers determined the reason for the above symptoms and the rapid spread of cases being a novel coronavirus named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), causative agent of the Coronavirus Disease-2019 (COVID-19)[2-4].
On 30 January 2020, World Health Organization (WHO) stated the novel coronavirus outburst in Wuhan, China, a global crisis[5]. Later on WHO accepted that SARS-CoV-2 has the ability to spread worldwide[6,7]. On 11 March 2020, the WHO announced COVID-19, a pandemic[8]. In successive months, several thousand people in different provinces of China and cities were invaded by the unchecked spread out of this disease[9]. Later, this disease traveled to various countries i.e. Thailand, Japan, Republic of Korea, Vietnam, Germany, United States, Singapore, and India. On comparison COVID-19 cases have overtaken the infected cases and deaths from Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and Severe Acute Respiratory Syndrome (SARS) at this point of the disease outburst[10]. The early effect of COVID-19 was so dreadful that the various countries had to implement phases of lockdowns. All age groups including children and pregnant women were badly affected due to this infectious disease.
CoVs (Coronaviruses) relates to the order Nidovirales and they have the largest RNA genome[11]. CoVs pertain to Coronaviridae family. They are positive single-stranded RNA-enveloped viruses. Four genera of CoVs are Alpha-, Beta-, Gamma-, and Deltacoronavirus. Seven human coronaviruses (HCoVs) have been revealed till now and they belong to the Alpha- and Betacoronavirus genera. The Alphacoronavirus genus includes HCoVNL63 and HCoV-229E and Betacoronavirus genus includes HCoV-OC43, HCoV-HKU1, SARS-CoV, MERS-CoV, and the novel SARS-CoV-2[12-17]. The alphacoronaviruses (HCoV-NL63 and HCoV-229E) and the betacoronaviruses (HCoV-OC43 and HCoV-HKU1) generally induce common colds, but severe lower respiratory tract infections can also appear, notably in the old age persons and kids[18]. HCoV-NL63 infection causes croup (laryngotracheitis)[19,20], and HCoV-OC43 infection causes severe lower respiratory tract infection among kids[21]. SARS-CoV and MERS-CoV are zoonotic viruses that cause severe respiratory syndrome[11].
This review summarizes the latest findings on the history, origin, transmission, genome structure, replication, epidemiology, pathogenesis, clinical features, diagnosis, and cure of COVID-19.
Human coronaviruses (229E and OC43) was first diagnosed in late 1960 as a reason for the common cold and were observed safe for human beings[22,23]. In Guangdong province in China in 2002–2003, a disease outbreak resulted in which a new coronavirus (β genera) originated in bats and was crossed to human beings by intermediate host of Himalayan palm civet cats[24]. This virus named SARS-CoV had a fatality rate of 10%[14,25,26]. This virus had been quickly spreading worldwide, particularly in Asia[27].
Almost ten years after SARS in year 2012, another highly pathogenic CoV, MERS-CoV, appeared in Middle East countries[17]. MERS-CoV, was also of bat origin, with dromedary camels as the intermediate host, and intermediate host reservoir species were also observed in goats, sheep, and cows[28]. MERS-CoV affected approximately 2000 people with approximately 34% mortality rate[17].
Recently, in December 2019, the novel Coronavirus 2019 (nCoV) or SARS-CoV-2 surfaced in Huanan Seafood Market, Wuhan (China) which cause pneumonia epidemic of unknown cause[29].
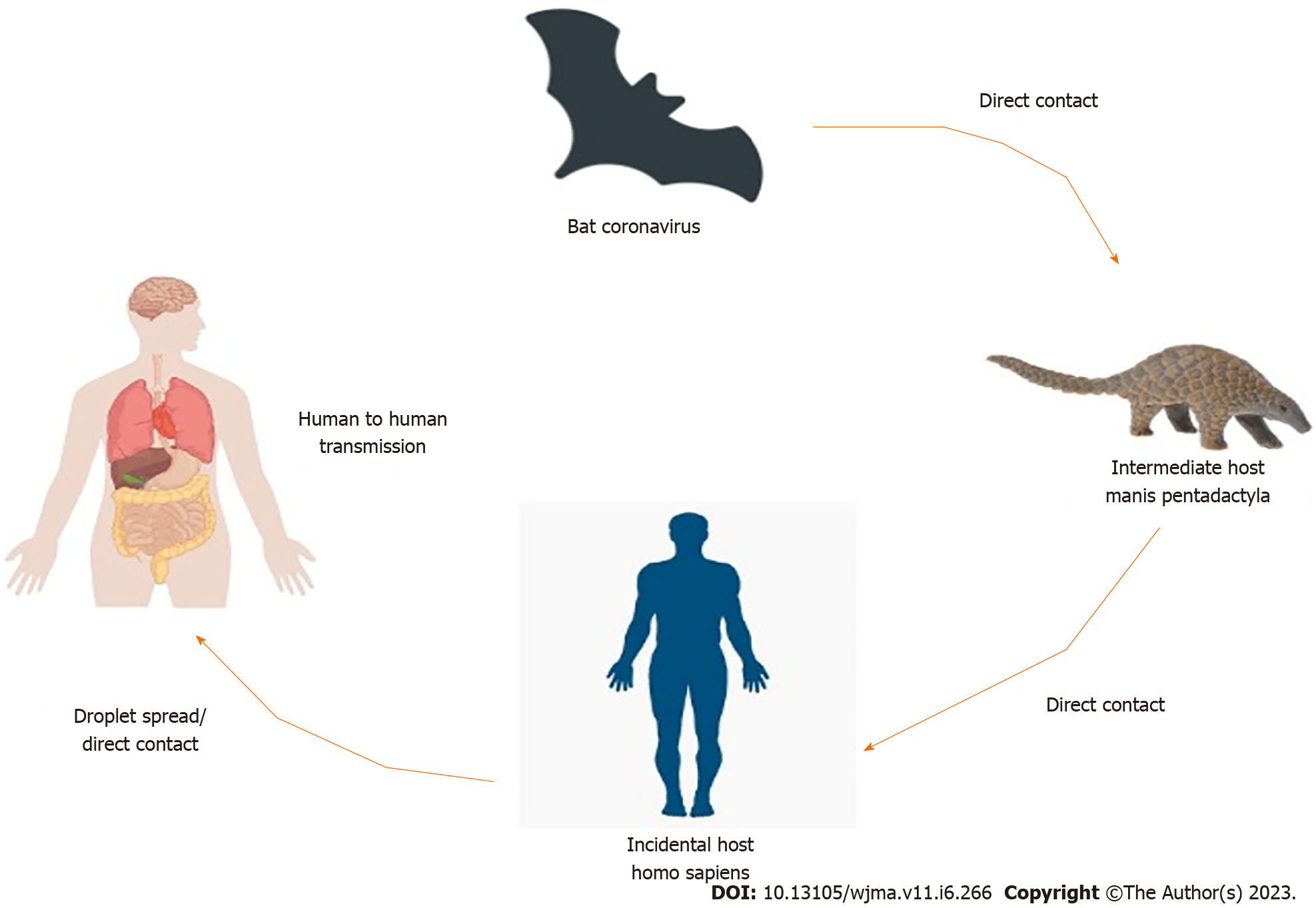
COVID-19 was thought to be originated in Wuhan (China). Environment specimens from the Huanan seafood market in Wuhan, China were examined positive, suggesting that the COVID-19 virus originated there[30]. According to several reports, Bat might be the likely pool of SARS-CoV-2[31,32]. Bats are the natural pool of a range of CoVs, including SARS-CoV-like and MERS-CoV-like viruses[33-35]. When the genome of COVID-19 and Bat CoV RaTG13 was compared and analyzed by virus genome sequencing and it revealed 96.2% genome sequence similarity with the Bat CoV RaTG13 genome[24]. It revealed that bat CoV and human SARS-CoV-2 might share the same ancestor[36]. It had > 70% resemblance with the SARS-CoV[37]. The SARS-CoV-2 emanated from bats and intermediate animals through which it reaches humans is unknown. Present suspects are pangolins and snakes[37]. Figure 1 shows the transmission cycle of SARS-CoV-2.
It seems that majority of early COVID-19 cases had a contact record with the seafood market, in Wuhan, China[24,38]. There is the possibility of human-to-human (Transmission via Aerosols, Nosocomial-Related Infections & Maternal Transmission) spread in people who did not have vulnerable to the seafood market of Wuhan, China[39]. It is also revealed that 31.3% of COVID-19 patients have traveled a short time ago to Wuhan and 72.3% of patients who are nonresidents of Wuhan, have contact with people of Wuhan[40]. Instances of COVID-19 in different provinces of China and in almost all countries of the world were recorded in people who were returning from Wuhan City, China[37]. COVID-19 cases were observed in countries outside China with no travel history to China indicating human-to-human transmission locally[41].
In India during the early period (from March 2020 onwards), there was an alarming rise in COVID-19 patients but now the recovery rate from this disease is much more and the situation is under control now.
SARS-CoV-2 belongs to beta-coronaviruses. Genome of SARS-CoV-2 is positive-sense single-stranded RNA [(+) ssRNA] with a 5'-cap, 3'-UTR poly(A) tail. The SARS-CoV-2 genome length is < 30 kb, having 14 open reading frames (ORFs) which encode non-structural proteins (NSPs), structural proteins i.e. spike (S), envelope (E), membrane/matrix (M) and nucleocapsid (N), and accessory proteins[42,43].
Coronavirus virions have a diameter of about 125 nm and are spherically shaped[44,45]. The genomes of coronaviruses encode five structural proteins: The spike (S), membrane (M), envelope (E) glycoproteins, hemagglutinin esterase (HE), and nucleocapsid (N) protein. All virions have all envelope protein and N protein, but only some beta coronaviruses possess the protein hemagglutinin esterase (HE)[46].
These proteins are located outside the virion and contribute to its usual shape. The homotrimers of the S proteins create the sun-like appearance that assigns coronaviruses their name[44,47,48]. Through their C-terminal transmembrane domains, S proteins attach to the virion membrane and also join with M proteins[49]. Virion attachment to particular surface receptors present in the host cell's plasma membrane is made possible by the N-terminus of the S proteins[50].
Three transmembrane domains are present in M glycoproteins. Glycosylation of M proteins occurs in the Golgi body[51-53]. Alteration in M protein is required to enter virion into the cell and for protein to become antigenic[54-56]. The M protein aids to regenerate new virions.
These are tiny proteins and are made from about 76 to 109 amino acids. The N-terminus of the E proteins typically has 30 amino acids, which facilitates adhesion to the virus membrane[57]. Additionally, Coronavirus E proteins perform an essential part in the assembly and morphogenesis of virions inside the cell.
They are phosphoproteins in nature. They possess flexible viral genomic RNA and have the ability to bind to helixes. N proteins perform a vital part in coronavirus virion structure, replication, and transcription[58,59].
The complete genome of Wuhan-Hu-1 coronavirus, a strain of SARS-CoV-2 (taken from a COVID-19 pneumonia patient), is of 29.9 kb size[36]. The CoVs genome contains between 6 and 11 ORFs[60]. Two polyproteins named pp1a and pp1ab, encode 16 non-structural proteins, which are translated by approximately 66% of the viral RNA present in the first ORF (ORF1a/b). The remaining ORFs form structural and accessory proteins. Spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein[11] are the four structural proteins encoded by the remaining part of the virus genome. SARS-CoV-2 is found to be more similar to SARS-like bat CoVs when compared with the known SARS-CoV and MERS-CoV genomes. The majority of genome-encoded proteins of SARS-CoV-2 are alike to those of SARS-CoVs. Zhang et al[61] observed that SARS-CoV-2 had been mutated in various patients in China. Tang et al[62] categorized two strains of SARS-CoV-2, the L type, and the S type. The L-type strains (derived from S-type) are more infectious and dangerous in terms of evolution than the S-type. As a result, virologists and epidemiologists must carefully examine the novel coronavirus and conduct additional research to determine its virulence and pandemic.
Here, we summarise the main steps of the SARS-CoV-2 infection cycle.
The human lower respiratory tract has ACE2, the SARS-CoV receptor[63]. Coronavirus S-glycoprotein may bind to ACE2 receptor present on outer surface of human cells[64]. S glycoprotein comprises of S1 and S2 subunits[65]. S1 subunit specifies the virus-host range and cellular tropism with the help of the RBD domain, whereas S2 subunit helps the fusion of virus with cell membrane with the help of heptad repeats 1 (HR1)[66] and heptad repeats 2 (HR2)[67] domains.
After fusing with the membrane, genomic RNA of virus is delivered inside the cytoplasm. This RNA forms pp1a and pp1ab polyproteins after translation[68], which further form non-structural proteins, and replication-transcription complex (RTC) in two-layered vesicles[69]. RTC replicates repeatedly and forms a set of subgenomic RNAs[70], which further form accessory proteins and structural proteins. Newly generated genomic RNA, nucleocapsids, and envelope glycoproteins unite to form new viral particles in the ER and Golgi apparatus[71].
At last, virion-containing vesicles combine along with the plasma membrane, and viruses are released outside.
This infection can affect people of any age. In humans, it is very contagious, especially in the elders and those who already have illnesses like fever, cold, or cough[72,73]. Large droplets released by symptomatic patients when coughing and sneezing are used to spread the infection; however, this can also happen from asymptomatic individuals prior to the start of symptoms[44]. COVID-19 infection transmits mainly by way of respiratory droplets, respiratory secretions, and direct contact[38]. Further, SARS-CoV-2 was also observed in faeces of severe pneumonia patients. Even after patients have recovered from the sickness, patients with symptoms can still spread infections. The infected droplets can deposit on surfaces and spread infection up to 1-2 meters away. In a suitable atmosphere, the virus can survive on surfaces for days. Disinfectants like hydrogen peroxide and sodium hypochlorite can destroy viruses[74]. Infection can be gained by inhaling infectious droplets or by touching surfaces that have been exposed to the virus and subsequently contacting mouth, nose, and eyes. Further, virus is found in faeces and affects the water reservoirs and then spreads by faeco-oral route or through aerosolization[75]. Transplacental transfer from pregnant women to their foetuses has not yet been documented. Although, post-natal transmission in neonates is reported[76]. The incubation period of this virus ranges from 2 to 14 d[77].
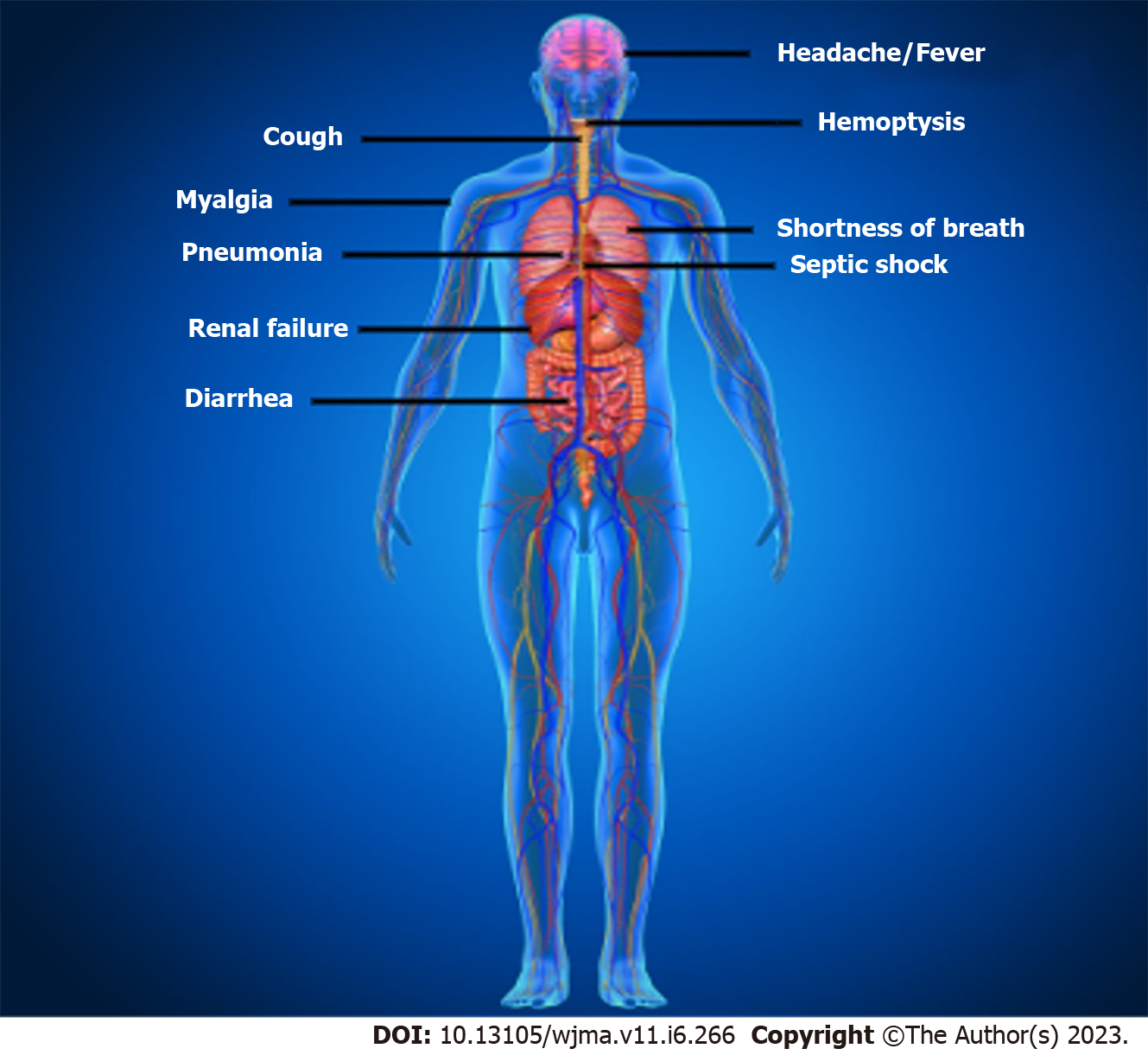
The clinical characteristics of patients with COVID-19 are shown in Figure 2. Asymptomatic state, acute respiratory distress syndrome, and multi-organ failure are all possible clinical manifestations of COVID-19[37]. Fever, coughing, sore throat, headaches, sputum production, sore throat, lethargy, myalgia, shortness of breath, and conjunctivitis are frequent clinical symptoms[37]. Acute respiratory distress syndrome (ARDS), arrhythmia, shock[78], acute renal injury, acute cardiac injury, liver dysfunction, and secondary infection were the disorders related to this infection[40]. This infection can lead to pneumonia, respiratory failure, and even death after the first week. IL2, IL7, IL10, GCSF, IP10, MCP1, MIP1A, and TNFα are inflammatory cytokines that have dramatically increased during the advancement of this disease[79]. Recovery from this infection began in the second or third week. Elderly persons are more likely to experience negative effects which can lead to death[37]. Additionally, it has been noted that this disease in neonates, kids, and children is substantially less severe than in adults[37].
An individual having fever, sore throat, and cough who has a traveling record to China or different locations with chronic community transmittance, or has contacted individuals having the same traveling experiences, or who have come into contact with a confirmed COVID-19 infected person is considered a suspected COVID-19 case[72]. A confirmed COVID-19 case is a suspected one having a positive molecular diagnostic test[72].
Until recently, the standard clinical diagnosis approach for COVID-19 is nucleic acid identification in swabs taken from nose, throat, or other parts of the respiratory system by using real-time polymerase chain reaction and furthermore verified by sequencing[80].
COVID-19 patients are adequately isolated to stop infection to other persons in contact, patients, and health personnel. Keeping adequate water in the body and a proper diet plan while managing fever and cough are the best ways to treat moderate infection at home. It is advised to provide oxygen to hypoxic patients using nasal prongs, face masks, a high-flow nasal cannula, or non-invasive ventilation[72].
Four classes of medicines have been identified based on how they work: (1) Viral entry and membrane fusion inhibitors; (2) protease inhibitors; (3) RdRp inhibitors; and (4) immunomodulatory medicines.
Table 1 shows various therapeutic agents used for the treatment of COVID-19.
| Sr. No. | Therapeutic agents | Examples |
| 1 | Antiviral agents | 1 Remdesivir |
| 2 Favipiravir | ||
| 3 Ribavirin | ||
| 4 Interferons | ||
| 5 Ritonavir/Lopinavir | ||
| 6 Arbidol | ||
| 7 Chloroquine/Hydroxychloroquine | ||
| 8 Recombinant soluble ACE2 | ||
| 9 Azithromycin | ||
| 10 Ivermectin | ||
| 11 Nitazoxanide | ||
| 12 Camostat mesylate | ||
| 13 Paxlovid | ||
| 2 | Biologic agents | 1 Monoclonal antibodies |
| 2 Convalescent plasma | ||
| 3 Hyperimmune sera | ||
| 4 Exogenous surfactant delivery | ||
| 3 | Anti-inflammatory agents | 1 Corticosteroids |
| 2 Fluvoxamine | ||
| 3 Anakinra | ||
| 4 Granulocyte-macrophage colony-stimulating factor inhibitors | ||
| 5 Intravenous immunoglobulin | ||
| 6 Janus kinase inhibitors | ||
| 7 Colchicine | ||
| 4 | Herbal agents | Various Chinese herbal medicine |
| 5 | Preventive agents | Vaccines |
Umifenovir, camostat mesylate, ACE inhibitors, angiotensin receptor-1 blockers, soluble recombinant human ACE2, chloroquine phosphate, and hydroxychloroquine sulfate are the various medications that were tested to prevent attachment and fusion of the virus to the cell membrane[81]. Due to their increased production capacity and lower danger of antibody-dependent enhancement, MAbs act more efficient than convalescent plasma as medication for COVID-19 patients[82]. A new MAb cocktail called REGN-COV2 binds to the receptor-binding domain of S1 or S2 subunits of the SARS-CoV-2 spike protein to stop the virus from entering the host cell[83]. Three more MAbs (B38, H4, and CR3022), might be potent against SARS-CoV-2 in upcoming studies[84,85].
Another class of medications that have been used for a long time to treat AIDS is protease inhibitors. Under the trade name Kaletra®, lopinavir is commonly compounded with ritonavir (LPV/r). The LPV/r effectiveness has been demonstrated earlier in cell culture as opposed to SARS-CoV-1 and MERS-CoV[86] and in recent times opposed to SARS-CoV-2[87].
RdRp inhibitors, in particular, demonstrated encouraging results in COVID-19 patients[88-90]. For instance, Remdesivir (RDV, GS-5734, Gilead) inhibited the spread of SARS-CoV-2 at smaller doses[89]. Another RdRp inhibitor, favipiravir (T-705, Avigan®), has demonstrated efficacy against SARS-CoV-2 in Vero E6 cells at higher concentrations[89]. Another RdRp inhibitors, such as β-D-N4-hydroxycytidine (EIDD-1931), were very effective at stopping SARS-CoV-1, SARS-CoV-2, and MERS-CoV replication in in vitro condition[91].
To lessen the intensity and complexities of COVID-19 and escape the inflammatory immune reactions (in serious patients), a variety of therapy is frequently applied[92]. Proinflammatory cytokine-suppressing medications, including MAbs (tocilizumab and sarilumab) and IL receptor inhibitors (anakinra), are now available[93]. In Vero E6 cells, nitazoxanide showed antiviral activity as opposed to SARS-CoV-2[89].
Corticosteroids aid to escape ARDS and acute lung injury by lowering cytokine storm and lung inflammation[94]. Induced pluripotent stem cells, mesenchymal stromal cells, and T cells are various cell therapy techniques that have been researched[95-98].
Currently, only a few approved medications are available to treat COVID-19 infection. Preventive measures play an important role to prevent this infection. It is advisable to keep confirmed or suspected cases having mild sickness isolated at home. Patients should wear a face mask and follow cough hygiene. Additionally, caregivers need to wash their hands regularly and should wear a surgical mask in the patient ward. Frequent sanitization of the rooms, surfaces, and equipment should be done with sodium hypochlorite. N95 respirators, safety suits, and goggles should be provided to healthcare professionals and workers. Healthcare professionals should also be frequently checked for various signs of COVID-19. Once a patient has been apyretic for at least three days and has two successive negative molecular tests with a sample gap of one day, they could be discharged from isolation. The only requirement for discharge was not the results of negative molecular tests[72].
Community-wide precautions include avoiding crowded places, forbidding large-scale gatherings, and delaying unnecessary travel to locations where transmission is still occurring. People should inculcate habit of good hand hygiene frequently, and exercise good cough hygiene by coughing into their sleeves or tissue paper rather than in their hands[99].
A law of banning the sale and trade of wild animals is also being introduced in China[100].
In this review, we outline the history, origin, transmission, genome structure, replication, epidemiology, pathogenesis, clinical characteristics, diagnosis, and treatment of COVID-19. The COVID-19 disease propagates rapidly across China and has disseminated to different countries of the world. Due to this viral epidemic, the economic, clinical, and public health frameworks of almost all countries of the world had affected. We wish that the horrible scenario created by this pandemic will not affect our life further.
The authors would like to thank Sh. Sanjay Kaushik, Lecturer (English), Government Model Sanskriti Senior Secondary School, Chulkana, Panipat, Haryana, India for timely support.
| 1. | Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J Med Virol. 2020;92:401-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1670] [Cited by in RCA: 1780] [Article Influence: 296.7] [Reference Citation Analysis (1)] |
| 2. | Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18987] [Cited by in RCA: 17892] [Article Influence: 2982.0] [Reference Citation Analysis (2)] |
| 3. | The New York Times. Coronavirus Updates: The Illness Now Has a Name, COVID-19. (accessed on 17 May 2020). Available from: https://www.nytimes.com/2020/02/11/world/asia/coronavirus-china.html. |
| 4. | Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470-473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4848] [Cited by in RCA: 4440] [Article Influence: 740.0] [Reference Citation Analysis (1)] |
| 5. | World Health Organization. International Health Regulations Emergency Committee on Novel Coronavirus in China. (accessed on 17 May 2020). Available from: https://www.who.int/news-room/events/detail/2020/01/30/default-calendar/international-health-regulationsemergency-committee-on-novel-coronavirus-in-china. |
| 6. | Business Insider. The Coronavirus Has Pandemic 'Potential' as it Spreads in South Korea, Italy, and Iran, According to WHO. (accessed on 7 April 2020). Available from: https://www.businessinsider.com/covid-19-coronavirus-has-pandemic-potential-says-who-2020-2?IR=T. |
| 7. | New Scientist. The WHO Still isn't Describing Covid-19 as a Pandemic. (accessed on May 17, 2020). Available from: https://www.newscientist.com/article/2235095-the-who-still-isnt-describing-covid-19-as-a-pandemic/#ixzz6F2fq8ncn. |
| 8. | World Health Organization. WHO Director-General's Opening Remarks at the Media Briefing on COVID-19-11 March 2020. (accessed on 17 May 2020). Available from: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020. |
| 9. | World Health Organization. Novel Coronavirus (2019-nCoV). Accessed February 7, 2020. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019. |
| 10. | Park M, Thwaites RS, Openshaw PJM. COVID-19: lessons from SARS and MERS. Eur J Immunol. 2020;50:308-11. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 11. | Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3747] [Cited by in RCA: 3406] [Article Influence: 486.6] [Reference Citation Analysis (1)] |
| 12. | Almeida JD, Tyrrell DA. The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J Gen Virol. 1967;1:175-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 146] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Kapikian AZ, James HD Jr, Kelly SJ, Dees JH, Turner HC, McIntosh K, Kim HW, Parrott RH, Vincent MM, Chanock RM. Isolation from man of "avian infectious bronchitis virus-like" viruses (coronaviruses) similar to 229E virus, with some epidemiological observations. J Infect Dis. 1969;119:282-290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 51] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10:S88-S97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 805] [Cited by in RCA: 747] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 15. | van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. Identification of a new human coronavirus. Nat Med. 2004;10:368-373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1250] [Cited by in RCA: 1312] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 16. | Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1037] [Cited by in RCA: 1118] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 17. | Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4030] [Cited by in RCA: 4093] [Article Influence: 292.4] [Reference Citation Analysis (0)] |
| 18. | Wevers BA, van der Hoek L. Recently discovered human coronaviruses. Clin Lab Med. 2009;29:715-724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | van der Hoek L, Sure K, Ihorst G, Stang A, Pyrc K, Jebbink MF, Petersen G, Forster J, Berkhout B, Uberla K. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005;2:e240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 218] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 20. | Choi EH, Lee HJ, Kim SJ, Eun BW, Kim NH, Lee JA, Lee JH, Song EK, Kim SH, Park JY, Sung JY. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000-2005. Clin Infect Dis. 2006;43:585-592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 281] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 21. | Zhu Y, Li C, Chen L, Xu B, Zhou Y, Cao L, Shang Y, Fu Z, Chen A, Deng L, Bao Y, Sun Y, Ning L, Liu C, Yin J, Xie Z, Shen K. A novel human coronavirus OC43 genotype detected in mainland China. Emerg Microbes Infect. 2018;7:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3085] [Cited by in RCA: 2935] [Article Influence: 489.2] [Reference Citation Analysis (0)] |
| 23. | Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605-614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 292] [Cited by in RCA: 306] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 24. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14339] [Article Influence: 2389.8] [Reference Citation Analysis (10)] |
| 25. | Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631-637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3643] [Cited by in RCA: 4193] [Article Influence: 190.6] [Reference Citation Analysis (1)] |
| 26. | Stadler K, Rappuoli R. SARS: understanding the virus and development of rational therapy. Curr Mol Med. 2005;5:677-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 27. | Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 691] [Cited by in RCA: 692] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 28. | Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 629] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 29. | Leroy EM, Ar Gouilh M, Brugère-Picoux J. The risk of SARS-CoV-2 transmission to pets and other wild and domestic animals strongly mandates a one-health strategy to control the COVID-19 pandemic. One Health. 2020;10:100133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 138] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 30. | Xinhua. China's CDC detects a large number of new coronaviruses in the South China seafood market in Wuhan, 2020. [RCA] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 31. | Giovanetti M, Benvenuto D, Angeletti S, Ciccozzi M. The first two cases of 2019-nCoV in Italy: Where they come from? J Med Virol. 2020;92:518-521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 224] [Cited by in RCA: 202] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 32. | Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 473] [Cited by in RCA: 414] [Article Influence: 69.0] [Reference Citation Analysis (0)] |
| 33. | Hampton T. Bats may be SARS reservoir. JAMA. 2005;294:2291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 34. | Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K. Bats and Coronaviruses. Viruses. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 297] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 35. | Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1723] [Cited by in RCA: 1821] [Article Influence: 86.7] [Reference Citation Analysis (0)] |
| 36. | Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6893] [Cited by in RCA: 7625] [Article Influence: 1270.8] [Reference Citation Analysis (1)] |
| 37. | Singhal T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J Pediatr. 2020;87:281-286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2006] [Cited by in RCA: 1591] [Article Influence: 265.2] [Reference Citation Analysis (0)] |
| 38. | Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382:1199-1207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11224] [Cited by in RCA: 9402] [Article Influence: 1567.0] [Reference Citation Analysis (0)] |
| 39. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30467] [Article Influence: 5077.8] [Reference Citation Analysis (13)] |
| 40. | Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS; China Medical Treatment Expert Group for Covid-19. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708-1720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19202] [Cited by in RCA: 19019] [Article Influence: 3169.8] [Reference Citation Analysis (9)] |
| 41. | Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M, Drosten C, Vollmar P, Zwirglmaier K, Zange S, Wölfel R, Hoelscher M. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382:970-971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2799] [Cited by in RCA: 2501] [Article Influence: 416.8] [Reference Citation Analysis (0)] |
| 42. | Abduljalil JM, Abduljalil BM. Epidemiology, genome, and clinical features of the pandemic SARS-CoV-2: a recent view. New Microbes New Infect. 2020;35:100672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (1)] |
| 43. | Parsamanesh N, Pezeshgi A, Hemmati M, Jameshorani M, Saboory E. Neurological manifestations of coronavirus infections: role of angiotensin-converting enzyme 2 in COVID-19. Int J Neurosci. 2022;132:917-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Bárcena M, Oostergetel GT, Bartelink W, Faas FG, Verkleij A, Rottier PJ, Koster AJ, Bosch BJ. Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc Natl Acad Sci U S A. 2009;106:582-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 199] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 45. | Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, Milligan RA, Yeager M, Buchmeier MJ. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol. 2006;80:7918-7928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 272] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 46. | Lissenberg A, Vrolijk MM, van Vliet AL, Langereis MA, de Groot-Mijnes JD, Rottier PJ, de Groot RJ. Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro. J Virol. 2005;79:15054-15063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 42] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 47. | Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84:3134-3146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 517] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 48. | Tan YJ, Lim SG, Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65:69-78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 79] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 49. | Chinese SARS Molecular Epidemiology Consortium. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666-1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 611] [Cited by in RCA: 552] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 50. | Lewicki DN, Gallagher TM. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J Biol Chem. 2002;277:19727-19734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 54] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | de Haan CA, Kuo L, Masters PS, Vennema H, Rottier PJ. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72:6838-6850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 207] [Article Influence: 7.4] [Reference Citation Analysis (14)] |
| 52. | Holmes KV, Doller EW, Sturman LS. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology. 1981;115:334-344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 144] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 53. | Niemann H, Geyer R, Klenk HD, Linder D, Stirm S, Wirth M. The carbohydrates of mouse hepatitis virus (MHV) A59: structures of the O-glycosidically linked oligosaccharides of glycoprotein E1. EMBO J. 1984;3:665-670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 50] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 54. | de Haan CA, de Wit M, Kuo L, Montalto-Morrison C, Haagmans BL, Weiss SR, Masters PS, Rottier PJ. The glycosylation status of the murine hepatitis coronavirus M protein affects the interferogenic capacity of the virus in vitro and its ability to replicate in the liver but not the brain. Virology. 2003;312:395-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 61] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Alexander S, Elder JH. Carbohydrate dramatically influences immune reactivity of antisera to viral glycoprotein antigens. Science. 1984;226:1328-1330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 107] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 56. | Wissink EHJ, Kroese MV, Maneschijn-Bonsing JG, Meulenberg JJM, van Rijn PA, Rijsewijk FAM, Rottier PJM. Significance of the oligosaccharides of the porcine reproductive and respiratory syndrome virus glycoproteins GP2a and GP5 for infectious virus production. J Gen Virol. 2004;85:3715-3723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 57. | Raamsman MJ, Locker JK, de Hooge A, de Vries AA, Griffiths G, Vennema H, Rottier PJ. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J Virol. 2000;74:2333-2342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 131] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 58. | Tok TT, Tatar G. Structures and Functions of Coronavirus Proteins: Molecular Modeling of Viral Nucleoprotein. Int J Virol Infect Dis. 2017;2:001-007. [DOI] [Full Text] |
| 59. | Weiss SR, Leibowitz JL. Coronavirus pathogenesis. Adv Virus Res. 2011;81:85-164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 590] [Cited by in RCA: 534] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 60. | Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, Zhu H, Zhao W, Han Y, Qin C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 688] [Cited by in RCA: 712] [Article Influence: 101.7] [Reference Citation Analysis (0)] |
| 61. | Zhang L, Shen FM, Chen F, Lin Z. Origin and Evolution of the 2019 Novel Coronavirus. Clin Infect Dis. 2020;71:882-883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 62. | Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z, Cui J, Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020;7:1012-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1089] [Cited by in RCA: 874] [Article Influence: 145.7] [Reference Citation Analysis (0)] |
| 63. | Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, McCray PB Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79:14614-14621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 620] [Cited by in RCA: 650] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 64. | Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 507] [Cited by in RCA: 570] [Article Influence: 81.4] [Reference Citation Analysis (0)] |
| 65. | Zhang N, Jiang S, Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev Vaccines. 2014;13:761-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 116] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 66. | Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, Ying T, Liu S, Shi Z, Jiang S, Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17:765-767. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 512] [Article Influence: 85.3] [Reference Citation Analysis (0)] |
| 67. | Yu F, Du L, Ojcius DM, Pan C, Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020;22:74-79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 68. | de Wilde AH, Snijder EJ, Kikkert M, van Hemert MJ. Host Factors in Coronavirus Replication. Curr Top Microbiol Immunol. 2018;419:1-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 248] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 69. | Sawicki SG, Sawicki DL. Coronavirus transcription: a perspective. Curr Top Microbiol Immunol. 2005;287:31-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 108] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 70. | Hussain S, Pan J, Chen Y, Yang Y, Xu J, Peng Y, Wu Y, Li Z, Zhu Y, Tien P, Guo D. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J Virol. 2005;79:5288-5295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 172] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 71. | Perrier A, Bonnin A, Desmarets L, Danneels A, Goffard A, Rouillé Y, Dubuisson J, Belouzard S. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J Biol Chem. 2019;294:14406-14421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 72. | Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, Fan YP, Fang C, Huang D, Huang LQ, Huang Q, Han Y, Hu B, Hu F, Li BH, Li YR, Liang K, Lin LK, Luo LS, Ma J, Ma LL, Peng ZY, Pan YB, Pan ZY, Ren XQ, Sun HM, Wang Y, Wang YY, Weng H, Wei CJ, Wu DF, Xia J, Xiong Y, Xu HB, Yao XM, Yuan YF, Ye TS, Zhang XC, Zhang YW, Zhang YG, Zhang HM, Zhao Y, Zhao MJ, Zi H, Zeng XT, Wang XH; , for the Zhongnan Hospital of Wuhan University Novel Coronavirus Management and Research Team, Evidence-Based Medicine Chapter of China International Exchange and Promotive Association for Medical and Health Care (CPAM). A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil Med Res. 2020;7:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 881] [Cited by in RCA: 1141] [Article Influence: 190.2] [Reference Citation Analysis (1)] |
| 73. | Poutanen SM, Low DE, Henry B, Finkelstein S, Rose D, Green K, Tellier R, Draker R, Adachi D, Ayers M, Chan AK, Skowronski DM, Salit I, Simor AE, Slutsky AS, Doyle PW, Krajden M, Petric M, Brunham RC, McGeer AJ; National Microbiology Laboratory, Canada; Canadian Severe Acute Respiratory Syndrome Study Team. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995-2005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 788] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 74. | Kampf G, Todt D, Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104:246-251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2034] [Cited by in RCA: 1994] [Article Influence: 332.3] [Reference Citation Analysis (0)] |
| 75. | World Health Organization. Situation reports. Accessed 22 Feb 2020. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. |
| 76. | Chen H, Guo J, Wang C, Luo F, Yu X, Zhang W, Li J, Zhao D, Xu D, Gong Q, Liao J, Yang H, Hou W, Zhang Y. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809-815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2400] [Cited by in RCA: 2278] [Article Influence: 379.7] [Reference Citation Analysis (1)] |
| 77. | Cheng ZJ, Shan J. 2019 Novel coronavirus: where we are and what we know. Infection. 2020;48:155-163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 339] [Cited by in RCA: 310] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 78. | Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14113] [Cited by in RCA: 14867] [Article Influence: 2477.8] [Reference Citation Analysis (1)] |
| 79. | Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14869] [Cited by in RCA: 13058] [Article Influence: 2176.3] [Reference Citation Analysis (4)] |
| 80. | Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1854] [Cited by in RCA: 2029] [Article Influence: 338.2] [Reference Citation Analysis (0)] |
| 81. | Hassanein SS, Sharaby MR, Tawfik NM, Rashed SA, Adel M, Fayez A, Mansour H, Amer HM. Latest Insights on the Diagnostic Approaches and Treatment Strategies of COVID-19. Intervirology. 2022;65:167-180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 82. | Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643-650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1091] [Cited by in RCA: 977] [Article Influence: 162.8] [Reference Citation Analysis (20)] |
| 83. | Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD; Trial Investigators. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N Engl J Med. 2021;384:238-251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1348] [Cited by in RCA: 1314] [Article Influence: 262.8] [Reference Citation Analysis (12)] |
| 84. | Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382-385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 899] [Cited by in RCA: 950] [Article Influence: 158.3] [Reference Citation Analysis (0)] |
| 85. | Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE, Tissot Dupont H, Honoré S, Colson P, Chabrière E, La Scola B, Rolain JM, Brouqui P, Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56:105949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3279] [Cited by in RCA: 3280] [Article Influence: 546.7] [Reference Citation Analysis (1)] |
| 86. | Chu CM, Cheng VC, Hung IF, Wong MM, Chan KH, Chan KS, Kao RY, Poon LL, Wong CL, Guan Y, Peiris JS, Yuen KY; HKU/UCH SARS Study Group. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1080] [Cited by in RCA: 1117] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 87. | Yamamoto N, Matsuyama S, Hoshino T, Yamamoto N. Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. bioRxiv. 2020. [DOI] [Full Text] |
| 88. | Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK; Washington State 2019-nCoV Case Investigation Team. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382:929-936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4155] [Cited by in RCA: 3853] [Article Influence: 642.2] [Reference Citation Analysis (2)] |
| 89. | Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269-271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4289] [Cited by in RCA: 4616] [Article Influence: 769.3] [Reference Citation Analysis (0)] |
| 90. | Williamson BN, Feldmann F, Schwarz B, Meade-White K, Porter DP, Schulz J, van Doremalen N, Leighton I, Yinda CK, Pérez-Pérez L, Okumura A, Lovaglio J, Hanley PW, Saturday G, Bosio CM, Anzick S, Barbian K, Cihlar T, Martens C, Scott DP, Munster VJ, de Wit E. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 478] [Cited by in RCA: 545] [Article Influence: 90.8] [Reference Citation Analysis (0)] |
| 91. | Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, Leist SR, Schäfer A, Dinnon KH 3rd, Stevens LJ, Chappell JD, Lu X, Hughes TM, George AS, Hill CS, Montgomery SA, Brown AJ, Bluemling GR, Natchus MG, Saindane M, Kolykhalov AA, Painter G, Harcourt J, Tamin A, Thornburg NJ, Swanstrom R, Denison MR, Baric RS. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 856] [Cited by in RCA: 827] [Article Influence: 137.8] [Reference Citation Analysis (0)] |
| 92. | Felsenstein S, Herbert JA, McNamara PS, Hedrich CM. COVID-19: Immunology and treatment options. Clin Immunol. 2020;215:108448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 428] [Cited by in RCA: 412] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 93. | Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, Diao K, Lin B, Zhu X, Li K, Li S, Shan H, Jacobi A, Chung M. Chest CT Findings in Coronavirus Disease-19 (COVID-19): Relationship to Duration of Infection. Radiology. 2020;295:200463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1728] [Cited by in RCA: 1607] [Article Influence: 267.8] [Reference Citation Analysis (3)] |
| 94. | Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;323:1824-1836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 978] [Cited by in RCA: 1279] [Article Influence: 213.2] [Reference Citation Analysis (0)] |
| 95. | Zaki MM, Lesha E, Said K, Kiaee K, Robinson-McCarthy L, George H, Hanna A, Appleton E, Liu S, Ng AHM, Khoshakhlagh P, Church GM. Cell therapy strategies for COVID-19: Current approaches and potential applications. Sci Adv. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 96. | Feins S, Kong W, Williams EF, Milone MC, Fraietta JA. An introduction to chimeric antigen receptor (CAR) T-cell immunotherapy for human cancer. Am J Hematol. 2019;94:S3-S9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 392] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 97. | Singh VK, Kalsan M, Kumar N, Saini A, Chandra R. Induced pluripotent stem cells: applications in regenerative medicine, disease modeling, and drug discovery. Front Cell Dev Biol. 2015;3:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 271] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 98. | Parekkadan B, Milwid JM. Mesenchymal stem cells as therapeutics. Annu Rev Biomed Eng. 2010;12:87-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 583] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 99. | World Health Organization. Coronavirus disease [COVID-19] Technical Guidance: Infection Prevention and Control. Accessed 20 Feb 2020. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/infection-prevention-and-control. |
| 100. | Li J, Li JJ, Xie X, Cai X, Huang J, Tian X, Zhu H. Game consumption and the 2019 novel coronavirus. Lancet Infect Dis. 2020;20:275-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Virology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D, D
Grade E (Poor): 0
P-Reviewer: Moreno-Galarraga L, Spain; Shen F, China; Sultana N, Bangladesh S-Editor: Liu JH L-Editor: A P-Editor: Yu HG