Published online Jun 26, 2014. doi: 10.5662/wjm.v4.i2.59
Revised: January 23, 2014
Accepted: March 17, 2014
Published online: June 26, 2014
Processing time: 265 Days and 18.8 Hours
MicroRNAs have become recognized as key players in the development of cancer. They are a family of small non-coding RNAs that can negatively regulate the expression of cancer-related genes by sequence-selective targeting of mRNAs, leading to either mRNA degradation or translational repression. Lung cancer is the leading cause of cancer-related death worldwide with a substantially low survival rate. MicroRNAs have been confirmed to play roles in lung cancer development, epithelial-mesenchymal transition and response to therapy. They are also being studied for their future use as diagnostic and prognostic biomarkers and as potential therapeutic targets. In this review we focus on the role of dysregulated microRNA expression in lung tumorigenesis. We also discuss the role of microRNAs in therapeutic resistance and as biomarkers. We further look into the progress made and challenges remaining in using microRNAs for therapy in lung cancer.
Core tip: Lung cancer is a prolific and high mortality disease, with few effective treatments. MicroRNAs have a role in the biogenesis and maintenance of lung cancer, with oncogenic and tumor suppressive effects. They are also a significant factor in resistance to current forms of therapy. There is evidence that microRNAs will be useful as diagnostic and predictive biomarkers in the future and, if delivery challenges can be overcome, they may become integrated into treatments.
- Citation: Joshi P, Middleton J, Jeon YJ, Garofalo M. MicroRNAs in lung cancer. World J Methodol 2014; 4(2): 59-72
- URL: https://www.wjgnet.com/2222-0682/full/v4/i2/59.htm
- DOI: https://dx.doi.org/10.5662/wjm.v4.i2.59
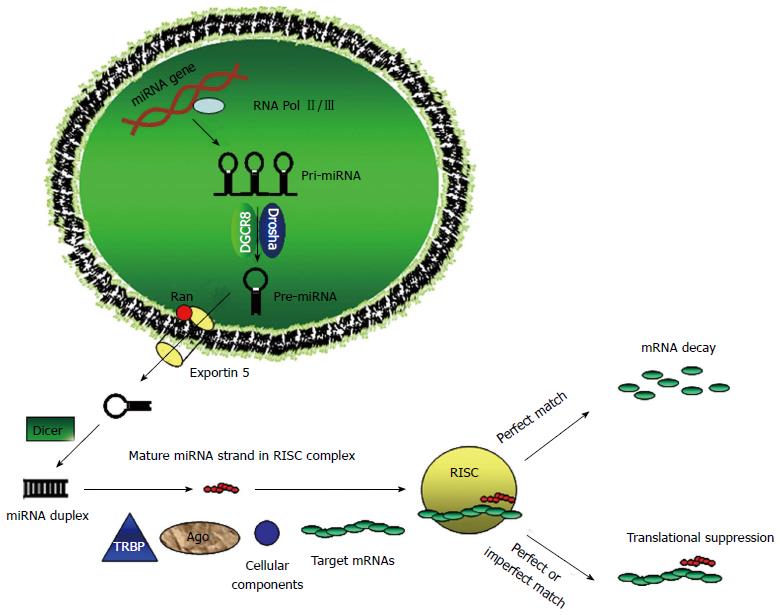
MicroRNAs (miRNAs) are small (19-22 nucleotides) non-coding RNAs that were first discovered in Caenorhabditis elegans[1]. MiRNAs silence their target genes by binding to the 3’ untranslated region (3’-UTR) of target messenger RNAs (mRNAs), causing either degradation or inhibition of translation. In animals, miRNAs are part of an approximately 70-100 nucleotides RNA with a stem-loop structure, known as a pre-miRNA that is included in hundreds or thousands of nucleotides long primary miRNA precursors (pri-miRNAs). The first step of microRNA biogenesis involves the transcription of the pri-miRNA and this is mediated by RNA polymerase II ( Pol-II)[2], although a minor group of microRNAs can be transcribed by RNA polymerase III (Pol-III)[3]. Then the pri-miRNA is processed in the nucleus by the RNase III enzyme Drosha and the protein Pasha/DGCR8 into pre-miRNAs[4]. The pre-miRNA undergoes a second processing step within the cytoplasm, and a small double-stranded RNA structure approximately 22 nucleotides in length is excised from the pre-miRNA hairpin by another RNase III enzyme, Dicer[5,6]. Finally, the mature single-stranded microRNA is loaded into the RNA-induced silencing complex, which mediates the degradation or translation inhibition of target mRNA by binding to its seed sequence in the target mRNA’s 3’-UTR (Figure 1). Dysfunctional microRNAs are commonly found in a variety of solid cancers and are attractive candidates for next-generation therapeutics.
Lung cancer remains the leading cause of cancer-related death worldwide, and non-small cell lung cancer (NSCLC) accounts for approximately 80% of all cases[7,8]. Although novel therapies targeting early diagnosis have been developed, the 5-year survival rate for NSCLC patients remains at a low 15%[9]. Takamizawa et al[10] were the first to relate microRNA expression to lung cancer. Since then there have been large number of studies relating microRNA expression with lung cancer. Here we describe the roles of microRNAs as tumor suppressors and oncogenes and their role in prognosis and diagnosis of lung cancer. Moreover, we discuss the contribution of microRNAs in radioresistance and chemoresistance as well as several therapeutic ventures involving microRNAs in lung cancer.
Numerous studies have reported finding mutation or aberrant expression of microRNAs in lung cancer patients. Investigators have shown that microRNAs whose expression is altered in tumors, may function as a novel class of oncogenes or tumor inhibitor genes. Several microRNAs are dysregulated in lung cancer, target cancer-relevant targets and have been documented to have tumor-suppressing or tumor-promoting activity in in vitro and/or in vivo models in lung cancer.
Let-7 was the first microRNA found to be dysregulated in lung cancer. Indeed, Takamizawa et al[10] reported that let-7 expression levels are frequently reduced in lung cancers both in vitro and in vivo. Let-7 subsequently was reported to be inversely correlated with RAS protein expression in lung cancer tissues, providing a possible mechanism for let-7 in lung cancer[11]. Kumar et al[12] used both inducible and constitutive expression systems to show substantial tumor suppression by let-7g in xenografts and a mouse lung tumor model in a K-Ras dependent manner. Enforced expression of let-7a in A549 cells decreased NIRF (Np95/ICBP90-like RING finger protein) leading to a coordinated increase in p21WAF1[13]. NIRF binds with higher affinity to the methylated CpGs of the promoter region through its SRA (SET and RING finger associated) domain and possibly recruits histone deacytylase-1 (HDAC1) through the same domain. This recruitment of HDAC1 to the methylated promoter regions of some tumor suppressor genes such as p21WAF1 can suppress their expression[14]. Recently[15], let-7c was observed to be inversely correlated to and directly target ITGB3 (integrin b3, also known as CD61)[16], and MAP4K3, a member of the MAP4K family[17] in NSCLC tissues. These observations support the assumption that let-7 may act as a tumor suppressor microRNA.
MiR-126 overexpression in lung cancer cell lines decreases Crk protein and leads to decreased adhesion, migration and invasion[18]. Crk is an adaptor protein that mediates several intracellular signal pathways[19] that are important in cell growth, motility, differentiation, and adhesion[20]. Liu et al[21] used an RNA protection assay to show downregulation of miR-126 in many lung cancer cell lines. MiR-126 overexpression efficiently reduced the expression of vascular endothelial growth factor (VEGF) and inhibited cell proliferation in vitro and tumorigenicity in vivo. Futhermore, enforced expression of miR-126 impaired NSCLC cell proliferation and tumor growth in xenografts model by targeting PIK3R2 and thus regulating the PI3K-Akt pathway[22], confirming a tumor suppressive role in lung cancer.
Introduction of miR-145 was reported to dramatically suppress the c-Myc/eIF4E pathway by targeting c-Myc, which has been demonstrated to be crucial for cell proliferation in NSCLC cells. Cell growth was inhibited and the G1/S transition was blocked by miR-145 overexpression in A549 and H23 cells[23]. Enforced expression of miR-145 negatively regulated the expression of EGFR and NUDT1[24]. NUDTI (8-oxo-dGTPase) is involved in accumulated mis-incorporation of oxidized 8-oxo-dGTP into DNA that can lead to cell dysfunction and death[25,26]. When miR-145 was overexpressed in A549 cell line there was a reduction in proliferation of CD133+ lung adenocarcinoma-initiating cells and tumorosphere growth capacity. This tumor suppressive effect involved miR-145 targeting of octamer-binding protein 4, a transcription factor of embryonic stem cells[27,28].
miR-200c plays a central role in the process of epithelial-mesenchymal transition (EMT) in highly invasive/aggressive NSCLC cells by targeting TCF8 (ZEB1) thus restoring its regulatory target E-cadherin[29,30]. Loss of miR-200c in invasive cells was observed to be a result of hypermethylation of the promoter region[30]. Studies by Yang et al[31] reveal a novel Jagged2/miR-200-dependent pathway that mediates lung adenocarcinoma EMT and metastasis in mice. They showed that Jagged2 increased the expression of GATA-binding factors that in turn suppressed members of miR-200 family driving EMT and reciprocally, miR-200 inhibited GATA3 expression reversing EMT. Furthermore, overexpression of miR-200 in murine lung adenocarcinoma cells decreased their growth and metastasis by targeting Flt1/VEGFR1[32] confirming the EMT suppressive function of miR-200.
The miR-34 family (miR-34a, -34b and -34c) is directly regulated by p53 and has been reported to induce apoptosis and cell cycle arrest in cancer cells[33,34] and is being studied for its anti-tumorigenic nature. The receptor tyrosine kinase Axl protein induces proliferation, migration and invasion in cancer[35]. Mudduluru et al[36,37] found an inverse correlation between Axl and miR-34a in NSCLC cell lines. ZEB1, a transcriptional repressor that promotes metastasis by downregulating microRNAs like the miR-200 family[38], drives prometastic actin cytoskeletal remodeling in NSCLC cells by inhibiting miR-34a expression[39]. Exogenous miR-34 prevented tumor initiation and progression in a therapeutically resistant KrasG12D/+; Trp53R172H/+ mouse lung cancer model[40]. Studies in our lab have shown that miR-34a and miR-34c overexpression increased TNF-related apoptosis inducing ligand (TRAIL)-induced apoptosis and decreased invasiveness of lung cancer cells by targeting PDGFR-α and PDGFR-β[41].
The miR-17-92 intronic cluster comprising seven different microRNAs namely miR-17-5p, -17-3p, -18a, -19a, -19b-1, -20a, and -92 was found by Hayashita et al[42] in lung cancer, mostly in small cell lung cancer (SCLC). Antisense oligonucleotides against mir-17-5p and miR-20a induced apoptosis in miR-17-92 overexpressing lung cancer cells[43]. Several targets have been studied for the various members of the miR-17-92 family. MiR-17-92 directly targeted hypoxia-inducible factor (HIF)-1A and overexpression of c-Myc led to downregulation of HIF-1A and induction of miR-17-92, suggesting that the induction of miR-17-92 may play a part in c-Myc–mediated repression of HIF-1A[44]. MiR-17-92 counterbalanced the generation of DNA damage in RB-inactivated SCLC cells by reducing γ-H2AX foci[45]. Matrix metalloproteinase (MMP) play an essential role in tumorigenesis by regulating migration and invasion of cells. In vivo MMP activity is controlled by the balance between MMPs and inhibitory proteins such as Reversion-inducing Cysteine-rich protein with Kazal motifs (RECK)[46]. STAT3 was shown to upregulate miR-92a thereby repressing RECK via post-transcriptional inhibition and thus promoting MMP activity[47]. These studies suggest that miR-17-92 may be an excellent therapeutic target candidate in the treatment of lung cancer.
miR-21 has been reported to be overexpressed in nine types of solid tumors including lung[48] as well as in hematological malignancies such as leukemia[49,50] and has great therapeutic potential for lung cancer. Over-expression of miR-21 enhanced tumorigenesis through inhibition of negative regulators of the Ras/ MEK/ERK pathway and inhibition of apoptosis[51]. MiR-21 was observed to repress phosphatase and tensin homolog (PTEN) and stimulated growth and invasion in NSCLC cell lines[52]. PTEN overexpression mimicked the same effects of anti-miR-21 such as inhibiting migration and invasion in NSCLC cells[53]. MiR-21 was shown to directly target the 3’-UTR of human mutS homolog 2, a core DNA mismatch repair (MMR) protein[54], thus affecting the cell cycle and cell proliferation in NSCLC cell lines[55] further underlining the oncogenic role of miR-21 in lung cancer.
MicroRNAs and DNA mismatch repair have been linked to human cancer progression. Human mutL homolog 1 (hMLH1) is a core MMR gene and reduced expression of hMLH1 can lead to genetic instability in NSCLC[56]. MiR-31-5p was reported to directly target and to be inversely correlated with MLH1 expression in NSCLC cell lines. Knockdown of miR-31-5p increased hMLH1 protein expression and induced a cell cycle arrest at G2/M phase in NSCLC cells[57]. MiR-31 was overexpressed in malignant lung tissues from humans and targeted the tumor-suppressive genes large tumor suppressor 2 (LATS2) and PP2A regulatory subunit B alpha isoform (PPP2R2A). Engineered knockdown of miR-31 repressed lung cancer cell growth and tumorigenicity in a dose-dependent manner[58]. These findings reveal that miR-31 acts as an oncogenic miRNA in lung cancer by targeting specific tumor suppressors for repression.
Our group has shown that miR-221 and -222 directly targeted PTEN and Tissue inhibitor of metalloproteinase 3 tumor suppressors inducing TRAIL resistance and enhancing cellular migration through the activation of the AKT pathway and MMPs. We observed that MET oncogene activates miR-221/222 through the c-Jun transcription factor[59]. The p53 up-regulated modulator of apoptosis (PUMA) suppresses growth of A549 cells through induction of apoptosis and sensitizes cells to chemotherapeutic agents and irradiation[60]. It was reported that miR-221/222 directly target and co-modulate PUMA expression and knockdown of miR-221/222 in A549 cells inhibited cell proliferation and induced mitochondrial-mediated apoptosis[61]. Thus, targeting miR-221/222 could be an effective strategy for therapy in lung cancer.
Radiotherapy, usually in combination with chemotherapy, is routinely used in lung cancer treatment, especially for NSCLC, allowing for better local control of the disease and reduction of metastasis occurrence. Both radiation resistance and chemoresistance is common, preventing successful long-term therapy and contributing to the dismal prognosis. Investigators are constantly trying to develop new effective therapies by studying the mechanisms behind resistance. Aberrant expression of several miRNAs has been correlated with the development and progression of tumors, and the reversal of their expression has been shown to modulate the cancer phenotype, suggesting the potential of miRNAs as targets for anti-cancer drugs. Here we describe the putative role(s) of microRNAs in the development of resistance to therapy (Table 1).
| miRNAs | Target | Drug/ Treatment | R/S | Ref. |
| miR-9, let-7g | NFκB1 | Radiotherapy | S | [66] |
| miR-34b | BCL2 | Radiotherapy | S | [67] |
| miR-214 | N/A; PTEN | Radiotherapy; gefitinib | R | [68,88] |
| miR-155 | FOXO3A; Apaf-1 | Radiotherapy; cisplatin | R | [69,78] |
| miR-210 | Stabilizes HIF-1A in normoxia | Radiotherapy | R | [70] |
| miR-181 | N/A | Cisplatin | S | [72] |
| miR-451 | N/A | Cisplatin | S | [73] |
| miR-98 | TP53 | Cisplatin | R | [74] |
| miR-497 | BCL2 | Multiple drugs | S | [75] |
| miR-200b | BCL2, XIAP; E2F3 | Cisplatin; docetaxel | S | [76,86] |
| miR-21 | PTEN, BCL2 | Cisplatin | R | [79] |
| miR-135a | APC | Paclitaxel | R | [82] |
| miR-100 | Plk1 | Docetaxel | S | [85] |
| miR-337-3p | STAT3, RAP1A | Paclitaxel and | S | [87] |
| docetaxel | ||||
| miR-221, miR-222 | P27kip1 | TRAIL | R | [91] |
| miR-130a | MET | TRAIL | S | [92] |
| miR-212 | PED | TRAIL | S | [93] |
When living cells are exposed to ionizing radiation (IR), a series of alterations occurs including transformation, cell cycle distress, mutations, sister-chromatid exchanges, chromosome aberrations, DNA repair, and apoptosis[62,63]. Among the IR-responsive genes, the activation of NFκB1 following genotoxic stress allows DNA damage repair and cell survival[64] and its inhibition can increase sensitivity of cancer cells to chemotherapeutic agents and radiation exposure[65]. Overexpression of miR-9 has been shown to down-regulate the level of NFκB1 in γ-irradiated H1299 human lung cancer cell line and decrease the surviving fraction of γ-irradiated cells. Interestingly, let-7g also suppressed the expression of NFκB1, although there is no canonical target site for let-7g in the NFκB1 3’-UTR[66]. Tumor suppressor p53 is another key player of the complex DNA damage response activated in response to IR[63]. Overexpression of p53-regulated miR-34b[33,34] in p53 wild type A549 cells increased radiosensitivity at low doses of radiation and this effect was not observed in p53 null H1299 cells[67].
Several microRNAs are involved in inducing resistance to irradiation. MiR-214 was shown to be upregulated in radioresistant NSCLC cells relative to radiosensitive counterparts and its overexpression protected radiosensitive cells against RT-induced apoptosis[68]. Incubation of NSCLC cell lines in hypoxic environments was reported to induce miR-155 expression and decrease its target, FOXO3A, a tumor suppressive transcription factor that regulates cell cycle and apoptosis. These increased levels of miR-155 radioprotected lung cancer cells and vice versa[69]. In another study, enforced expression of miR-210 increased radioresistance of NSCLC cells by stabilizing HIF-1A[70]. These studies reveal a therapeutically important link between miRNA expression, hypoxia, and irradiation.
Platinum agents like cisplatin and carboplatin are some of the principal chemotherapeutic agents used for treatment of NSCLC. These agents induce their cytotoxic effects by targeting cellular DNA and are active against a number of tumour types[71]. However numerous studies have shown that an initial success associated with partial responses or disease stabilization is followed by the selection of chemotherapy-resistant tumor cells, leading to chemotherapeutic failure. Numerous microRNAs have been implicated in cisplatin resistance. Galluzi and colleagues reported miR-181a and miR-630 to be the most upregulated miRNAs after cisplatin (CDDP) treatment however, pre-miR-181a enhanced while pre-miR-630 reduced CDDP-triggered cell death in A549 cells by modulating steps of the intrinsic pathway of apoptosis[72]. Another group observed that ectopic expression of miR-451 might be involved in sensitizing A549 cells to cisplatin by inducing apoptosis via inactivation of Akt signaling pathway and enhancement of caspase-3 activity[73]. Zhang et al[74] showed that transfection with miR-98/miR-453 inhibited p53 expression and upon treatment with cisplatin, the expression of miR-98 decreases, while p53 increases. This led them to speculate that regulation of p53 pathway might play an important role in the action of cisplatin on A549 cell growth. Separate studies by Zhu et al[75,76] on miR-497 and miR-200b/429 cluster in multidrug resistant A549/CDDP cell line indicated an increased sensitivity to cisplatin in part by modulation of apoptosis via targeting only B-cell CLL/lymphoma 2 (BCL2) or both BCL2 and X-linked inhibitor of apoptosis, respectively. In the cytosol, Apaf-1 can bind with cytochrome-c released from the mitochondrial inter-membrane, and activate the initiator caspase-9, eventually resulting in cellular apoptosis[77]. MiR-155 was observed to be inversely correlated to Apaf-1 in lung cancer tissues. Silencing miR-155 or overexpressing Apaf-1 in A549 cell lines greatly increased the sensitivity of A549 cells to cisplatin treatment through an Apaf-1 mediated pathway, involving increased expression of Bax and caspase-9[78]. MiR-21 was reported to be critical in platinum resistance in NSCLC and modulated the sensitivity of NSCLC cells to platinum, at least in part, by regulating PTEN and BCL-2 expressions[79].
Taxanes, such as paclitaxel and docetaxel, are chemotherapeutic drugs that stabilize microtubules and inhibit their disassembly to tubulin interfering with proper formation of the mitotic spindle, which leads to activation of the mitotic spindle checkpoint and mitotic arrest[80]. Drug-treated cells then undergo apoptosis as a result of the abnormal mitosis[81]. Studies reporting the role of microRNAs in taxane resistance can provide novel adjuvant strategies along with taxanes in the treatment of lung cancer. Knockdown of miR-135a was reported to upregulate adenomatous polyposis coli gene (APC) and sensitize paclitaxel-resistant NSCLC cell lines to paclitaxel-induced cell death[82]. APC is a tumor suppressor that regulates the mitotic checkpoint by binding to microtubules during mitosis[83]. Polo-like kinase (Plk)-1 is a cell cycle protein that plays an important role in spindle dynamics and chromosome segregation during mitosis[84]. Feng et al[85] showed that introduction of miR-100 resensitized docetaxel resistant SPC-A1/DTX cells to docetaxel by suppression of cell proliferation, enhancement of apoptosis, and cell arrest in G2/M phase of cell cycle at least partially by Plk-1 targeting. The same group also reported that ectopic expression of miR-200b reversed docetaxel resistance of SPC-A1/DTX cells in part by targeting E2F3[86]. Du and colleagues identified a novel regulatory pathway involving STAT3 and RAP1A that modulates miR-337-3p mediated paclitaxel sensitivity in lung cancer cells[87].
Patients with NSCLC who have activating epidermal growth factor receptor (EGFR) mutations derive clinical benefit from treatment with EGFR-tyrosine kinase inhibitors (EGFR-TKIs)-namely gefitinib and erlotinib. However, these patients eventually develop resistance to EGFR-TKIs. Wang et al[88] established a gefitinib resistant cell line-HCC827/GR and found that miR-214 was significantly up-regulated in these cells compared to control HCC827 cells. Knockdown of miR-214 in HCC827/GR resulted in upregulation of PTEN and inactivation of p-AKT and this in turn re-sensitized the cells to gefitinib. To understand the role of microRNAs in TKI-resistant NSCLCs, our group examined miRNA dysregulation mediated by TK receptors. MiR-30b, -30c, -221 and -222 were found to be modulated by both EGFR and MET receptors whereas miR-103 and miR-203 were controlled only by MET. We showed that these miRNAs influenced the response to gefitinib of NSCLC cells in vitro and in vivo by inhibiting the expression of the genes encoding BCL2-like 11 (BIM), apoptotic peptidase activating factor 1 (APAF-1), protein kinase Cε (PKC-ε) and sarcoma viral oncogene homolog[89].
Treatment with TRAIL induces programmed cell death in a wide range of transformed cells, both in vitro and in vivo, without producing significant effects in normal cells[90]. However, a significant proportion of human cancer cells are resistant to TRAIL-induced apoptosis, and the mechanisms of sensitization vary among cell types. To define novel pathways that regulate TRAIL-sensitivity in NSCLC, our lab performed genome-wide expression profiling of microRNAs. Levels of miR-221 and -222 were increased in TRAIL-resistant NSCLC cells and their knockdown rendered CALU-1-resistant cells sensitive to TRAIL. Conversely, H460-sensitive cells treated with pre-miR-221 and -222 developed a resistance. Interference with TRAIL signaling by miR-221 and -222 was mainly through targeting p27kip1[91]. Another study from our lab further reported that miR-130a, expressed at low level in lung cancer cell lines, by targeting MET was able to reduce TRAIL resistance in NSCLC cells through the c-Jun-mediated downregulation of miR-221 and miR-222[92]. Ectopic expression of miR-212 increased TRAIL-induced cell death in NSCLC cells by targeting PED/PEA-15 (PED), a death effector domain family member with a broad anti-apoptotic function[93]. These studies enhance our understanding of the mechanisms responsible for TRAIL resistance.
The diagnosis of lung cancer is performed through several methods with varying degrees of sensitivity and reliability. X-ray imaging, along with positron emission tomography and computed tomography (CT) scans, is often the first diagnostic procedure utilized. While these methods provide valuable information when anomalies are easily visible, problems with lung segmentation and positioning in the chest cavity, human error and competent detection software prevent imaging from always producing successful diagnoses. Similarly, while tissue sampling though bronchoscopy has become the standard practice in diagnosing lung cancer, it presents its own difficulties, including complications in obtaining viable samples due to patient symptoms, proper imaging and tumor position[94].
MicroRNAs show potential as biomarkers for the diagnosis of lung cancer that can complement and improve upon other techniques. Promising lung cancer microRNA biomarkers can be found circulating in the bloodstream, in sputum and inside cells, and are detected at an abnormal level when cancer is present. Ideally, these biomarkers should be detected through minimally invasive methods and with limited discomfort to patients. There are currently several dozen microRNAs under investigation for their biomarker properties (Tables 2 and 3).
| Traditional Procedures | Possible microRNA biomarkers | |
| Diagnosis | ||
| Detect abnormalities | X-ray, CT scan | 21 |
| Confirm malignancy | Biopsy, sputum/fluid cytology | let-7, 29a, 34c, 205, 375 |
| Prognosis | ||
| Staging | CT scan, PET, MRI | 21, 125b, 155, 182/183 |
| Mutational status | Sequencing, PCR, microarray | 21, 155 |
| Treatment | Potential prognostic biomarkers | Potential role in resistance |
| Surgery | Let-7, 21 | |
| Radiotherapy | 155, 210 | Let-7g, 9, 34, 155, 210, 214 |
| Chemotherapy | 21, 125b | 21, 30b/c, 98, 100, 103, 130a, 135a, 155, 181, 200b, 203, 212, 214, 221/222, 337, 451, 453, 494, 630 |
MiR-21 has a well-documented correlation to lung cancer. Detected in both serum and sputum, elevated miR-21 corresponds to lowered survival rate, lymphoid invasions and KRAS mutations[53,95]. Promisingly, assays for miR-21 in sputum from lung cancer patients have shown higher sensitivity than traditional sputum cytology with very high specificity[96,97]. MiR-21, in combination with miR-210 and miR-486-5p, was shown to be expressed significantly higher in the plasma of patients with malignant solitary pulmonary nodules (SPNs) compared to those with benign SPNs. Solitary pulmonary nodules have been increasingly diagnosed with the improvement of CT scan technology and its widespread use. However, only a small fraction of SPNs are malignant. The combination of miR-21 testing and CT scans could provide a minimally invasive method of determining the cancer status of patients with SPNs[98].
MiR-155 is a prominent oncomiR, with various roles in lung cancer including proliferation and drug resistance. Used in a panel with miR-197 and miR-182, miR-155 was able to distinguish between NSCLC patients and control samples by real time PCR of plasma. Patients with metastasizing cancer consistently exhibited higher levels of plasma miR-155, which could additionally aid in staging the disease[99]. Several studies have found that miR-155 is only elevated in EGFR/KRAS-negative lung cancer. Samples from surgically resected lung specimens and fine needle aspirations (FNAs) both demonstrated this effect[100,101]. FNAs are considered to be safe, minor surgical procedures compared to excisional biopsies, thus further development of its use for collection of miR-155 to determine mutational status could be beneficial. An even less invasive technique was used by Yao and colleagues to determine levels of miR-155 in vivo[102]. They developed a novel molecular beacon that can be introduced into mice with lipid-DNA complexes and detect miR-155 in lung cancer xenografts through in vivo fluorescent imaging. The authors posit that these results may be translatable to human lung tumors, possibly improving on the problematic imaging resources currently available for diagnostics.
The miR-183 family (miR-96, miR-182, and miR-183) is a group of oncomiRs that have been confirmed to be overexpressed in lung tumors and serum in NSCLC. Targets for these miRs support a variety of biological processes, including growth, migration, invasion and angiogenesis. MiR-182 in particular has been found to be strongly correlated to primary tumors while all three are expressed more in squamous cell carcinoma than in adenocarcinoma[103-105]. Additionally, miR-182 showed a high specificity and sensitivity, and readily differentiates stage I lung cancer from normal control samples, making it a tantalizing possibility for non-invasive clinical diagnostics. MiR-183, on the other hand, has been demonstrated as being able to differentiate between early and late stage NSCLC, while not being able to discriminate early stage lung cancer from normal cells[106].
The miR-34 family, in particular miR-34a and miR-34c, has been shown by multiple groups to be potential biomarkers in lung cancer[100]. Mascaux and colleagues detailed an inverse relationship between miR-34 levels and lung carcinogenesis, and later expounded on this by showing that changes in lung cell histology are reflected by miR-34c independent of any treatment[107]. However, these studies used biopsies as their source tissue, and have not yet confirmed that other extraction methods (sputum, serum, etc.) could be used to circumvent normal tissue sampling and histology. Akbas et al[108] have found that dysregulation of 34c can be confirmed through serum in chronic obstructive pulmonary disease (COPD) - an inflammatory disease that increases the risk of lung cancer - making further development of 34c as a lung cancer biomarker a likely avenue of research.
NSCLC is the most common form of lung cancer, and is divided into two subtypes, squamous cell carcinoma (SCC) and adenocarcinoma. Diagnosing the correct subtype is critical for treatment and microRNA biomarkers that are able to distinguish between these subtypes would be a useful tool in a clinical setting. MiR-205 - a tumor suppressor - has been found by several researchers to be a highly effective identifier of squamous cell histology through its downregulation, both in NSCLC tissues and serum[109,110]. Similarly, members of the let-7 family are significantly downregulated in SCC, likely due to the fact that let-7 regulates RAS expression, and RAS mutations are far more common in SCC than adenocarcinomas[111,112]. As lung tissue has one of the highest expressions of let-7 in the body, its characteristic decrease in SCC has the potential to make it an easily identifiable biomarker[113]. However, because this large decrease in expression has only been identified in lung tissue and not sputum, serum or bronchial fluids, the only current options for assaying let-7 are tissue biopsy or bronchial brushing. Thus, let-7 as a biomarker will have to show greater efficacy than traditional cytopathology to warrant clinical use.
Though less common and with comparatively little research, small cell lung cancer presents serious problems for patients, with a tendency toward rapid and widespread metastasis. Therefore, accurate and expeditious diagnostic markers are desirable. Two studies have found miR-375 to accurately discriminate between NSCLC and SCLC. Huang et al[114] used snap-frozen and paraffin-embedded surgical lung specimens, finding miR-29a and miR-375 to be superior to traditional cytopathology for diagnosing SCLC. In the other study, Zhao and colleagues found extremely elevated miR-375 expression in four human SCLC cell lines and four SCLC-like cell lines generated in mice[115]. These results are promising, but require more study with larger sample populations and examinations of extracellular microRNA levels to evaluate the usefulness of miR-375 as a clinical biomarker.
An essential facet of cancer treatment involves the correct and efficient prognosis of the type of cancer and the expectations of survival and mortality. This prevents the unneeded use of potentially harmful drugs, and allows for the correct prescription of treatment strength and severity. As microRNAs have been confirmed to play roles in lung cancer development, migration and response to therapy, they may also find future use as biomarkers to give accurate prognoses to physicians.
Liu et al[116] found that miR-21 was significantly elevated in the serum of NSCLC patients with lower survival rates and showed a strong association with lymph node metastasis and advanced clinical stage. Yang et al[117] confirmed this result with a meta-analysis while others found similar results in three ethnically-diverse cohorts, including significant associations between elevated miR-21 and high-mortality stage I tumors[118,119]. These findings have the potential of allowing physicians to quickly evaluate and escalate treatments in response to early stage NSCLC diagnoses. Studies examining post-operative lung cancer patients also found that miR-21 serum levels significantly decreased in response to successful surgery, with higher miR-21 expression corresponding to shorter survival time and disease recurrence[120]. However, another study evaluating the use of miR-21 as a predictive biomarker in SCLC found no correlation between miR-21 expression and patient outcome[121]. The same study found similar results with 6 other important NSCLC-related miRs, underlying both the inherent differences between SCLC and NSCLC and also the paucity of data involving SCLC biomarkers. MiR-155 has been shown to have a similar elevation in expression in NSCLC, which is associated with low survival and high rates of recurrence[122,123]. Both miR-21 and miR-155 have been examined in sputum samples and found in readily detectable quantities, and while Xie and collaborators found only miR-21 produces adequate differentiation in expression for use as a biomarker[97], others have found using both in combination with three other miRs to be a highly sensitive panel for clinical applications[124].
The let-7 family, in addition to discriminating between SCC and adenocarcinoma, has also been found to be associated with survival rate. Low let-7a expression has been shown by multiple studies to correlate to a poor prognosis, both pre- and post-operative, particularly in SCC[125]. There is some evidence that this preferential prognostic ability comes from the squamous cell carcinoma’s reliance on the downregulation of tumor suppressor miRs, including let-7, compared to adeocarcimoma’s dependence on the upregulation of oncomiRs[111].
Several miRs have been identified as having the potential to predict the effectiveness of therapy. MiR-125b is an oncomiR that has been found to be significantly increased in stage III and IV NSCLC. Cui et al[126] examined the expression of a panel of miRs, finding that miR-125b levels were markedly higher in patients that did not respond to cisplatin treatment. This corroborates several other studies that found that miR-125b inhibited cisplatin-induced apoptosis in breast and ovarian cancers[127,128]. Similarly, miR-21 shows promise as predictive biomarker for the response to adjuvant platinum based chemotherapies (cisplatin, oxaliplatin, etc.) in NSCLC[79]. Serum taken from patients after surgery and platinum based treatment showed elevated levels of miR-21 compared to a pretreatment baseline if there was a low chemotherapeutic response. A recent study found that serum miR-210 consistently determined the success of platinum based chemotherapy. MiR-210 is upregulated in NSCLC and recent findings have shown that patients who responded well to treatment had significantly lower expression of miR-210 in serum, near levels expected in healthy control subjects[129]. Another study reported that NSCLC cells overexpressing miR-210 were conferred with radioresistance as well, displaying an ability to rapidly repair double-strand DNA breaks[70]. As radiotherapy is a common treatment in lung cancer, with more than half of patients receiving irradiation, potential microRNA biomarkers - miR-210 for example - that predict the efficacy of this procedure would make an immediate impact on patients and physicians’ decisions. In its role as a mediator of radioresistance, miR-155 may also have potential as a prognostic biomarker, with elevated expression corresponding to lower survival rate in patients who have received radiotherapy[69,130].
Increasing evidence supporting the essential role of microRNAs in the machinery of cancer points to the possibility of using microRNAs as treatments in lung cancer. The most evident problem blocking clinical use of microRNA therapies is delivery. Specifically targeting cancer cells, maintaining microRNA stability in bodily fluids and penetration of cellular membranes are areas of intense investigation. Some modifications to microRNA and anti-microRNA oligonucleotides (AMOs), including 2’-O-methyl, 2’-O-methoxyethyl and locked nucleic acids, provide nuclease resistance for greater longevity during serum transport, but additional methods are needed to enhance the cell permeability of these molecules[131].
Liposomes are a promising avenue of microRNA therapy delivery. These artificial, spherical vesicles made from a lipid bilayer are used to administer pharmaceutical drugs, microRNA or small interfering RNA (siRNA). Experiments using mouse models have found both neutral lipid emulsions and cationic lipoplexes to be effective in delivering microRNAs to lung tumors. Multiple studies have used a neutral lipid emulsion to deliver tumor suppressors miR-34a and let-7 to NSCLC tumors in mice, which resulted in a 60% reduction in tumor area[132,133]. Wu and colleagues found that cationic lipoplexes were over 50-fold more effective in delivering pre-miR-133b, a known inhibitor of NSCLC proliferation, to NSCLC mice than NeoFX complexes, a standard transfection reagent, and with lower cytotoxicity[134,135]. Recently, this same team used cationic lipoplexes to deliver miR-29b into murine A549 xenografts, finding similar success in cellular penetration, along with documenting a decrease in tumorigenicity and improved functionality of cisplatin[136]. Shi and colleagues have used a novel technology - solid lipid nanoparticles (SLNs) - to transport AMOs to suppress miR-21 in lung cancer and introduce miR-34a into lung cancer stem cells, inhibiting cell migration and inducing cell apoptosis[137]. SLNs boast superior cellular uptake rates and decreased oligonucleotide degradation, which allow AMOs to be introduced without stability-adding modifications that reduce specificity. Future directions for liposome therapy reseach include increasing stability of liposomes and better targeting through the use of tumor-recognizing antibodies and peptides. Some studies have already shown that incorporating ligands that target overexpressed lung cancer receptors into liposomes dramatically improves liposome uptake into NSCLC cells[138] while others have used synthetic antigens to activate tumor-targeting immune cells[139].
Viral delivery systems are a platform that offers naturally high infection rates and high miRNA expression levels for lung cancer treatment. Adenoviruses have been used as a vector for the delivery of miR-122, a tumor suppressor, into NSCLC NCI-H460 cells. The resulting 2000-fold higher expression of miR-122 led to the activation of intrinsic apoptotic pathways[140]. Sun and colleagues used a lentiviral vector to infect hepatocellular carcinoma cells with osteopontin-suppressing microRNAs that decreased tumorigenicity in mice and downregulated the oncogenic MEK/ERK/1/2 pathway[141]. These results may be transferable to lung cancer, as osteopontin has been identified as a pro-metastatic factor in NSCLC[142]. Overall, adenoviruses are considered the better option for microRNA vectors as they do not integrate into the genome. There are complications with viral delivery of microRNAs, though, including immunogenicity and cellular toxicity that will need to be addressed in further research.
As liposome delivery often produces toxicity and requires considerable optimization to maintain adequate stability and efficacy, and viral vectors are limited by immunogenicity much research has recently focused on the use of nanoparticles. These small, solid spheres offer reduced immune response, lower toxicity and cheap, efficient production methods that result in high complex stability.
Protamine, a biologically derived molecule, has been complexed with microRNA, resulting in higher transfection rates than with lipoplexes[143]. Chen et al[144] utilized protamine complexes with miR-34a to inhibit the growth of lung metastases of melanoma. These nanoparticles incorporated a liposome shell around the nanoparticles, with great effectiveness. Further studies are needed to determine the necessity of liposomal encirclement of protamine complexes, taking into account microRNA degradation, cellular uptake and immune response. Gold and silica nanoparticles have also been utilized in microRNA delivery[145,146], but as of yet, there are no studies demonstrating their use in treating lung cancer.
MiRNAs have become recognized as key players in cancer. Their ability to regulate expression of cancer-related genes has immense implications for the diagnosis and treatment of cancer. Lung cancer is the leading cause of cancer-related death worldwide and currently has a substantially lower survival rate than many other common cancers. In this review, we discuss how dysregulated miRNA expression has been shown to contribute to the genesis and maintenance of lung cancer, through the down-regulation of tumor suppressors and up-regulation of oncomiRs. Additionally, miRNAs may be essential in the development of chemo- and radioresistance in lung cancer. Due to their importance in the regulatory structure of cancer, miRNAs may soon be used to improve diagnosis and predictions of outcomes and response to therapy, although more studies will be needed with larger sample groups to resolve conflicting reports of disease-state expression patterns for some miRNAs. Implementing miRNAs and anti-miRNAs as treatments presents some additional difficulties, mostly related to delivery and stability inside the body, but holds promise as a less toxic therapy that can target multiple genes simultaneously. The investigation into miRNAs and cancer is still relatively new, and more study will be needed to form consensuses on the critical functions of miRNAs inside cancer cells, what information can be gleaned from changes in their expression and the best methods for therapeutic administration, but these unique compounds show great promise as tools against lung cancer.
| 1. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [PubMed] |
| 2. | Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-4060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2902] [Cited by in RCA: 3042] [Article Influence: 138.3] [Reference Citation Analysis (0)] |
| 3. | Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 943] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 4. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3513] [Cited by in RCA: 3675] [Article Influence: 159.8] [Reference Citation Analysis (0)] |
| 5. | Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1990] [Cited by in RCA: 2006] [Article Influence: 80.2] [Reference Citation Analysis (0)] |
| 6. | Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654-2659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1305] [Cited by in RCA: 1338] [Article Influence: 53.5] [Reference Citation Analysis (11)] |
| 7. | Ramalingam S, Pawlish K, Gadgeel S, Demers R, Kalemkerian GP. Lung cancer in young patients: analysis of a Surveillance, Epidemiology, and End Results database. J Clin Oncol. 1998;16:651-657. [PubMed] |
| 8. | Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7953] [Cited by in RCA: 8116] [Article Influence: 477.4] [Reference Citation Analysis (4)] |
| 9. | Miller YE. Pathogenesis of lung cancer: 100 year report. Am J Respir Cell Mol Biol. 2005;33:216-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 105] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753-3756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1813] [Cited by in RCA: 1853] [Article Influence: 84.2] [Reference Citation Analysis (0)] |
| 11. | Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2664] [Cited by in RCA: 2734] [Article Influence: 130.2] [Reference Citation Analysis (0)] |
| 12. | Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903-3908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 681] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 13. | He X, Duan C, Chen J, Ou-Yang X, Zhang Z, Li C, Peng H. Let-7a elevates p21(WAF1) levels by targeting of NIRF and suppresses the growth of A549 lung cancer cells. FEBS Lett. 2009;583:3501-3507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601-7610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 253] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 15. | Zhao B, Han H, Chen J, Zhang Z, Li S, Fang F, Zheng Q, Ma Y, Zhang J, Wu N. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 16. | Bennett JS, Catella-Lawson F, Rut AR, Vilaire G, Qi W, Kapoor SC, Murphy S, FitzGerald GA. Effect of the Pl(A2) alloantigen on the function of beta(3)-integrins in platelets. Blood. 2001;97:3093-3099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 52] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 17. | Diener K, Wang XS, Chen C, Meyer CF, Keesler G, Zukowski M, Tan TH, Yao Z. Activation of the c-Jun N-terminal kinase pathway by a novel protein kinase related to human germinal center kinase. Proc Natl Acad Sci USA. 1997;94:9687-9692. [PubMed] |
| 18. | Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, Nuovo G, Marsh CB, Nana-Sinkam SP. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373:607-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 221] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 19. | Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348-6371. [PubMed] |
| 20. | Kobashigawa Y, Tanaka S, Inagaki F. Structural basis for the transforming activity of human cancer-related signaling adaptor protein Crk. Tanpakushitsu Kakusan Koso. 2008;53:148-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 21. | Liu B, Peng XC, Zheng XL, Wang J, Qin YW. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 263] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 22. | Yang J, Lan H, Huang X, Liu B, Tong Y. MicroRNA-126 inhibits tumor cell growth and its expression level correlates with poor survival in non-small cell lung cancer patients. PLoS One. 2012;7:e42978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 23. | Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng A, Hu J. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J Exp Clin Cancer Res. 2010;29:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 202] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 24. | Cho WC, Chow AS, Au JS. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol. 2011;8:125-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 180] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 25. | Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J Biol Chem. 1993;268:23524-23530. [PubMed] |
| 26. | Yoshimura D, Sakumi K, Ohno M, Sakai Y, Furuichi M, Iwai S, Nakabeppu Y. An oxidized purine nucleoside triphosphatase, MTH1, suppresses cell death caused by oxidative stress. J Biol Chem. 2003;278:37965-37973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 115] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 27. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14584] [Article Influence: 810.2] [Reference Citation Analysis (0)] |
| 28. | Yin R, Zhang S, Wu Y, Fan X, Jiang F, Zhang Z, Feng D, Guo X, Xu L. microRNA-145 suppresses lung adenocarcinoma-initiating cell proliferation by targeting OCT4. Oncol Rep. 2011;25:1747-1754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972-7976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 328] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 30. | Ceppi P, Mudduluru G, Kumarswamy R, Rapa I, Scagliotti GV, Papotti M, Allgayer H. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res. 2010;8:1207-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 255] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 31. | Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, Alvarez CA, Moreira DC, Creighton CJ, Gregory PA. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121:1373-1385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 32. | Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbons DL, Baird BN, Alvarez C, Thilaganathan N, Liu DD, Saintigny P. miR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 2011;9:25-35. [PubMed] |
| 33. | He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130-1134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2135] [Cited by in RCA: 2119] [Article Influence: 111.5] [Reference Citation Analysis (0)] |
| 34. | Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 832] [Cited by in RCA: 876] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 35. | Vajkoczy P, Knyazev P, Kunkel A, Capelle HH, Behrndt S, von Tengg-Kobligk H, Kiessling F, Eichelsbacher U, Essig M, Read TA. Dominant-negative inhibition of the Axl receptor tyrosine kinase suppresses brain tumor cell growth and invasion and prolongs survival. Proc Natl Acad Sci USA. 2006;103:5799-5804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 201] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 36. | Mudduluru G, Allgayer H. The human receptor tyrosine kinase Axl gene--promoter characterization and regulation of constitutive expression by Sp1, Sp3 and CpG methylation. Biosci Rep. 2008;28:161-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 37. | Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene. 2011;30:2888-2899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 205] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 38. | Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 671] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 39. | Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, Pertsemlidis A, Gregory PA, Wright JA, Goodall GJ. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest. 2012;122:3170-3183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 40. | Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72:5576-5587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 210] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 41. | Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. MiR-34a/c-Dependent PDGFR-α/β Downregulation Inhibits Tumorigenesis and Enhances TRAIL-Induced Apoptosis in Lung Cancer. PLoS One. 2013;8:e67581. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 42. | Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628-9632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1174] [Cited by in RCA: 1215] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 43. | Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M, Nimura Y. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26:6099-6105. |
| 44. | Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68:5540-5545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 235] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 45. | Ebi H, Sato T, Sugito N, Hosono Y, Yatabe Y, Matsuyama Y, Yamaguchi T, Osada H, Suzuki M, Takahashi T. Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene. 2009;28:3371-3379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 85] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 46. | Noda M, Oh J, Takahashi R, Kondo S, Kitayama H, Takahashi C. RECK: a novel suppressor of malignancy linking oncogenic signaling to extracellular matrix remodeling. Cancer Metastasis Rev. 2003;22:167-175. [PubMed] |
| 47. | Lin HY, Chiang CH, Hung WC. STAT3 upregulates miR-92a to inhibit RECK expression and to promote invasiveness of lung cancer cells. Br J Cancer. 2013;109:731-738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 48. | Liu M, Jiang S, Lu Z, Young K, Li Y. Physiological and pathological functions of mammalian microRNAs. In McQueen CA. Comprehensive toxicology. Newnes. 2010;2:427-446. |
| 49. | Tagawa H, Ikeda S, Sawada K. Role of microRNA in the pathogenesis of malignant lymphoma. Cancer Sci. 2013;104:801-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 50. | Feng Y, Chen X, Gao L. Knockdown of miR-21 as a novel approach for leukemia therapy. J Formos Med Assoc. 2010;109:621-623. [PubMed] |
| 51. | Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282-293. [PubMed] [DOI] [Full Text] |
| 52. | Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta. 2010;411:846-852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 456] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 53. | Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372:35-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 263] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 54. | Jascur T, Boland CR. Structure and function of the components of the human DNA mismatch repair system. Int J Cancer. 2006;119:2030–2035. |
| 55. | Zhong Z, Dong Z, Yang L, Gong Z. miR-21 induces cell cycle at S phase and modulates cell proliferation by down-regulating hMSH2 in lung cancer. J Cancer Res Clin Oncol. 2012;138:1781-1788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 56. | Kouso H, Yoshino I, Miura N, Takenaka T, Ohba T, Yohena T, Osoegawa A, Shoji F, Maehara Y. Expression of mismatch repair proteins, hMLH1/hMSH2, in non-small cell lung cancer tissues and its clinical significance. J Surg Oncol. 2008;98:377-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 57. | Zhong Z, Dong Z, Yang L, Chen X, Gong Z. MicroRNA-31-5p modulates cell cycle by targeting human mutL homolog 1 in human cancer cells. Tumour Biol. 2013;34:1959-1965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 58. | Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298-1309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 59. | Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P. miR-221 & 222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498-509. [PubMed] [DOI] [Full Text] |
| 60. | Yu J, Yue W, Wu B, Zhang L. PUMA sensitizes lung cancer cells to chemotherapeutic agents and irradiation. Clin Cancer Res. 2006;12:2928-2936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 74] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 61. | Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, Pu P, Kang C. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol. 2010;37:1621-1626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 62. | Preston RJ. Radiation biology: concepts for radiation protection. Health Phys. 2005;88:545-556. [PubMed] |
| 63. | Amundson SA. Functional genomics in radiation biology: a gateway to cellular systems-level studies. Radiat Environ Biophys. 2008;47:25-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 64. | Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 275] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 65. | Li F, Sethi G. Targeting transcription factor NF-kappaB to overcome chemoresistance and radioresistance in cancer therapy. Biochim Biophys Acta. 2010;1805:167-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 222] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 66. | Arora H, Qureshi R, Jin S, Park AK, Park WY. miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFκB1. Exp Mol Med. 2011;43:298-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 91] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 67. | Balça-Silva J, Sousa Neves S, Gonçalves AC, Abrantes AM, Casalta-Lopes J, Botelho MF, Sarmento-Ribeiro AB, Silva HC. Effect of miR-34b overexpression on the radiosensitivity of non-small cell lung cancer cell lines. Anticancer Res. 2012;32:1603-1609. [PubMed] |
| 68. | Salim H, Akbar NS, Zong D, Vaculova AH, Lewensohn R, Moshfegh A, Viktorsson K, Zhivotovsky B. miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. Br J Cancer. 2012;107:1361-1373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 69. | Babar IA, Czochor J, Steinmetz A, Weidhaas JB, Glazer PM, Slack FJ. Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer Biol Ther. 2011;12:908-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 70. | Grosso S, Doyen J, Parks SK, Bertero T, Paye A, Cardinaud B, Gounon P, Lacas-Gervais S, Noël A, Pouysségur J. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013;4:e544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 190] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 71. | Go RS, Adjei AA. Review of the comparative pharmacology and clinical activity of cisplatin and carboplatin. J Clin Oncol. 1999;17:409–422. |
| 72. | Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupé P, Robert T. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793-1803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 225] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 73. | Bian HB, Pan X, Yang JS, Wang ZX, De W. Upregulation of microRNA-451 increases cisplatin sensitivity of non-small cell lung cancer cell line (A549). J Exp Clin Cancer Res. 2011;30:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 123] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 74. | Zhang S, Zhang C, Li Y, Wang P, Yue Z, Xie S. miR-98 regulates cisplatin-induced A549 cell death by inhibiting TP53 pathway. Biomed Pharmacother. 2011;65:436-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 75. | Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang B, Shu Y, Liu P. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2012;29:384-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 76. | Zhu W, Xu H, Zhu D, Zhi H, Wang T, Wang J, Jiang B, Shu Y, Liu P. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother Pharmacol. 2012;69:723-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 77. | Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 739] [Cited by in RCA: 803] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 78. | Zang YS, Zhong YF, Fang Z, Li B, An J. MiR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther. 2012;19:773-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 79. | Gao W, Lu X, Liu L, Xu J, Feng D, Shu Y. MiRNA-21: a biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther. 2012;13:330-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 115] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 80. | Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277:665-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2545] [Cited by in RCA: 2554] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 81. | Shi J, Orth JD, Mitchison T. Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res. 2008;68:3269-3276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 180] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 82. | Holleman A, Chung I, Olsen RR, Kwak B, Mizokami A, Saijo N, Parissenti A, Duan Z, Voest EE, Zetter BR. miR-135a contributes to paclitaxel resistance in tumor cells both in vitro and in vivo. Oncogene. 2011;30:4386-4398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 83. | Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es JH, Breukel C, Wiegant J, Giles RH. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol. 2001;3:433-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 507] [Cited by in RCA: 495] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 84. | Golsteyn RM, Mundt KE, Fry AM, Nigg EA. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J Cell Biol. 1995;129:1617-1628. [PubMed] |
| 85. | Feng B, Wang R, Chen LB. MiR-100 resensitizes docetaxel-resistant human lung adenocarcinoma cells (SPC-A1) to docetaxel by targeting Plk1. Cancer Lett. 2012;317:184-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 86. | Feng B, Wang R, Song HZ, Chen LB. MicroRNA-200b reverses chemoresistance of docetaxel-resistant human lung adenocarcinoma cells by targeting E2F3. Cancer. 2012;118:3365-3376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 107] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 87. | Du L, Subauste MC, DeSevo C, Zhao Z, Baker M, Borkowski R, Schageman JJ, Greer R, Yang CR, Suraokar M. miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS One. 2012;7:e39167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 88. | Wang YS, Wang YH, Xia HP, Zhou SW, Schmid-Bindert G, Zhou CC. MicroRNA-214 regulates the acquired resistance to gefitinib via the PTEN/AKT pathway in EGFR-mutant cell lines. Asian Pac J Cancer Prev. 2012;13:255-260. [PubMed] |
| 89. | Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2012;18:74-82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 314] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 90. | Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol. 2007;39:1462-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 350] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 91. | Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845-3855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 222] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 92. | Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli G. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 93. | Incoronato M, Garofalo M, Urso L, Romano G, Quintavalle C, Zanca C, Iaboni M, Nuovo G, Croce CM, Condorelli G. miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 2010;70:3638-3646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 107] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 94. | Reck M, Hermes A, Tan EH, Felip E, Klughammer B, Baselga J. Tissue sampling in lung cancer: a review in light of the MERIT experience. Lung Cancer. 2011;74:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 95. | Capodanno A, Boldrini L, Proietti A, Alì G, Pelliccioni S, Niccoli C, D’Incecco A, Cappuzzo F, Chella A, Lucchi M. Let-7g and miR-21 expression in non-small cell lung cancer: correlation with clinicopathological and molecular features. Int J Oncol. 2013;43:765-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 96. | Li Y, Li W, Ouyang Q, Hu S, Tang J. Detection of lung cancer with blood microRNA-21 expression levels in Chinese population. Oncol Lett. 2011;2:991-994. [PubMed] |
| 97. | Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 249] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 98. | Shen J, Liu Z, Todd NW, Zhang H, Liao J, Yu L, Guarnera MA, Li R, Cai L, Zhan M. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011;11:374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 217] [Cited by in RCA: 205] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 99. | Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, Wang MX. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4:575-586. [PubMed] |
| 100. | Dacic S, Kelly L, Shuai Y, Nikiforova MN. miRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Mod Pathol. 2010;23:1577-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 101. | Petriella D, Galetta D, Rubini V, Savino E, Paradiso A, Simone G, Tommasi S. Molecular profiling of thin-prep FNA samples in assisting clinical management of non-small-cell lung cancer. Mol Biotechnol. 2013;54:913-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 102. | Yao Q, Zhang AM, Ma H, Lin S, Wang XX, Sun JG, Chen ZT. Novel molecular beacons to monitor microRNAs in non-small-cell lung cancer. Mol Cell Probes. 2012;26:182-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 103. | Tan X, Qin W, Zhang L, Hang J, Li B, Zhang C, Wan J, Zhou F, Shao K, Sun Y. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res. 2011;17:6802-6811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 104. | Barshack I, Lithwick-Yanai G, Afek A, Rosenblatt K, Tabibian-Keissar H, Zepeniuk M, Cohen L, Dan H, Zion O, Strenov Y. MicroRNA expression differentiates between primary lung tumors and metastases to the lung. Pathol Res Pract. 2010;206:578-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 105. | Zhu W, Liu X, He J, Chen D, Hunag Y, Zhang YK. Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: a case control study. BMC Cancer. 2011;11:393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 106. | Lin Q, Mao W, Shu Y, Lin F, Liu S, Shen H, Gao W, Li S, Shen D. A cluster of specified microRNAs in peripheral blood as biomarkers for metastatic non-small-cell lung cancer by stem-loop RT-PCR. J Cancer Res Clin Oncol. 2012;138:85-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 107. | Mascaux C, Feser WJ, Lewis MT, Barón AE, Coldren CD, Merrick DT, Kennedy TC, Eckelberger JI, Rozeboom LM, Franklin WA. Endobronchial miRNAs as biomarkers in lung cancer chemoprevention. Cancer Prev Res (Phila). 2013;6:100-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 108. | Akbas F, Coskunpinar E, Aynaci E, Oltulu YM, Yildiz P. Analysis of serum micro-RNAs as potential biomarker in chronic obstructive pulmonary disease. Exp Lung Res. 2012;38:286-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 109. | Lu Y, Govindan R, Wang L, Liu PY, Goodgame B, Wen W, Sezhiyan A, Pfeifer J, Li YF, Hua X. MicroRNA profiling and prediction of recurrence/relapse-free survival in stage I lung cancer. Carcinogenesis. 2012;33:1046-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 110. | Zhang YK, Zhu WY, He JY, Chen DD, Huang YY, Le HB, Liu XG. miRNAs expression profiling to distinguish lung squamous-cell carcinoma from adenocarcinoma subtypes. J Cancer Res Clin Oncol. 2012;138:1641-1650. [PubMed] |
| 111. | Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola FM. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 265] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 112. | Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776-5783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 315] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 113. | Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713-7722. [PubMed] |
| 114. | Huang W, Hu J, Yang DW, Fan XT, Jin Y, Hou YY, Wang JP, Yuan YF, Tan YS, Zhu XZ. Two microRNA panels to discriminate three subtypes of lung carcinoma in bronchial brushing specimens. Am J Respir Crit Care Med. 2012;186:1160-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 115. | Zhao H, Zhu L, Jin Y, Ji H, Yan X, Zhu X. miR-375 is highly expressed and possibly transactivated by achaete-scute complex homolog 1 in small-cell lung cancer cells. Acta Biochim Biophys Sin (Shanghai). 2012;44:177-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 116. | Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, Zeng F, Zhou JH, Zhang YK. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 117. | Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong W, Liao Y, Du J. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer. 2013;49:604-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 183] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 118. | Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, Bowman ED, Mathé EA, Takenoshita S, Yokota J, Haugen A. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17:1875-1882. [PubMed] |
| 119. | Akagi I, Okayama H, Schetter AJ, Robles AI, Kohno T, Bowman ED, Kazandjian D, Welsh JA, Oue N, Saito M. Combination of protein coding and noncoding gene expression as a robust prognostic classifier in stage I lung adenocarcinoma. Cancer Res. 2013;73:3821-3832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 120. | Le HB, Zhu WY, Chen DD, He JY, Huang YY, Liu XG, Zhang YK. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol. 2012;29:3190-3197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 121. | Lee JH, Voortman J, Dingemans AM, Voeller DM, Pham T, Wang Y, Giaccone G. MicroRNA expression and clinical outcome of small cell lung cancer. PLoS One. 2011;6:e21300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 122. | Tang D, Shen Y, Wang M, Yang R, Wang Z, Sui A, Jiao W, Wang Y. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev. 2013;22:540-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 123. | Wang Y, Li J, Tong L, Zhang J, Zhai A, Xu K, Wei L, Chu M. The prognostic value of miR-21 and miR-155 in non-small-cell lung cancer: a meta-analysis. Jpn J Clin Oncol. 2013;43:813-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 124. | Roa WH, Kim JO, Razzak R, Du H, Guo L, Singh R, Gazala S, Ghosh S, Wong E, Joy AA. Sputum microRNA profiling: a novel approach for the early detection of non-small cell lung cancer. Clin Invest Med. 2012;35:E271. [PubMed] |
| 125. | Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189-198. [PubMed] |
| 126. | Cui EH, Li HJ, Hua F, Wang B, Mao W, Feng XR, Li JY, Wang X. Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta Pharmacol Sin. 2013;34:309-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 127. | Wang H, Tan G, Dong L, Cheng L, Li K, Wang Z, Luo H. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS One. 2012;7:e34210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 209] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 128. | Kong F, Sun C, Wang Z, Han L, Weng D, Lu Y, Chen G. miR-125b confers resistance of ovarian cancer cells to cisplatin by targeting pro-apoptotic Bcl-2 antagonist killer 1. J Huazhong Univ Sci Technolog Med Sci. 2011;31:543-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 129. | Li ZH, Zhang H, Yang ZG, Wen GQ, Cui YB, Shao GG. Prognostic significance of serum microRNA-210 levels in nonsmall-cell lung cancer. J Int Med Res. 2013;41:1437-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 130. | Chen J, Wang BC, Tang JH. Clinical significance of microRNA-155 expression in human breast cancer. J Surg Oncol. 2012;106:260-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 131. | Liu Z, Sall A, Yang D. MicroRNA: An emerging therapeutic target and intervention tool. Int J Mol Sci. 2008;9:978-999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 123] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 132. | Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923-5930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 543] [Cited by in RCA: 528] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 133. | Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, Weidhaas JB, Bader AG, Slack FJ. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116-1122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 553] [Cited by in RCA: 525] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 134. | Wu Y, Crawford M, Yu B, Mao Y, Nana-Sinkam SP, Lee LJ. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 2011;8:1381-1389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 135. | Liu L, Shao X, Gao W, Zhang Z, Liu P, Wang R, Huang P, Yin Y, Shu Y. MicroRNA-133b inhibits the growth of non-small-cell lung cancer by targeting the epidermal growth factor receptor. FEBS J. 2012;279:3800-3812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 136. | Wu Y, Crawford M, Mao Y, Lee RJ, Davis IC, Elton TS, Lee LJ, Nana-Sinkam SP. Therapeutic Delivery of MicroRNA-29b by Cationic Lipoplexes for Lung Cancer. Mol Ther Nucleic Acids. 2013;2:e84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 178] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 137. | Shi SJ, Zhong ZR, Liu J, Zhang ZR, Sun X, Gong T. Solid lipid nanoparticles loaded with anti-microRNA oligonucleotides (AMOs) for suppression of microRNA-21 functions in human lung cancer cells. Pharm Res. 2012;29:97-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 138. | Dubey PK, Singodia D, Vyas SP. Polymeric nanospheres modified with YIGSR peptide for tumor targeting. Drug Deliv. 2010;17:541-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 139. | Hansen J, Lindenstrøm T, Lindberg-Levin J, Aagaard C, Andersen P, Agger EM. CAF05: cationic liposomes that incorporate synthetic cord factor and poly(I: C) induce CTL immunity and reduce tumor burden in mice. Cancer Immunol Immunother. 2012;61:893-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 140. | Ma L, Liu J, Shen J, Liu L, Wu J, Li W, Luo J, Chen Q, Qian C. Expression of miR-122 mediated by adenoviral vector induces apoptosis and cell cycle arrest of cancer cells. Cancer Biol Ther. 2010;9:554-561. [PubMed] |
| 141. | Sun BS, Dong QZ, Ye QH, Sun HJ, Jia HL, Zhu XQ, Liu DY, Chen J, Xue Q, Zhou HJ. Lentiviral-mediated miRNA against osteopontin suppresses tumor growth and metastasis of human hepatocellular carcinoma. Hepatology. 2008;48:1834-1842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 143] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 142. | Wang XM, Li J, Yan MX, Liu L, Jia DS, Geng Q, Lin HC, He XH, Li JJ, Yao M. Integrative analyses identify osteopontin, LAMB3 and ITGB1 as critical pro-metastatic genes for lung cancer. PLoS One. 2013;8:e55714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 72] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 143. | Suh JS, Lee JY, Choi YS, Chung CP, Park YJ. Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomaterials. 2013;34:4347-4359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 144. | Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18:1650-1656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 404] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 145. | Ghosh R, Singh LC, Shohet JM, Gunaratne PH. A gold nanoparticle platform for the delivery of functional microRNAs into cancer cells. Biomaterials. 2013;34:807-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 146. | Tivnan A, Orr WS, Gubala V, Nooney R, Williams DE, McDonagh C, Prenter S, Harvey H, Domingo-Fernández R, Bray IM. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PLoS One. 2012;7:e38129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 184] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
P- Reviewer: Hattori N, Jayaprakash V S- Editor: Wen LL L- Editor: A E- Editor: Wu HL