Published online Jun 20, 2024. doi: 10.5662/wjm.v14.i2.92267
Revised: February 19, 2024
Accepted: April 12, 2024
Published online: June 20, 2024
Processing time: 145 Days and 17.6 Hours
Ocular surface squamous neoplasia (OSSN) is a common eye surface tumour, characterized by the growth of abnormal cells on the ocular surface. OSSN in
Core Tip: This is a unique comprehensive review written on a common eye tumour-ocular surface squamous neoplasia (OSSN) and the role of artificial intelligence (AI) in the early diagnosis and management of this ocular condition. This write-up also covers the various intricacies involved in developing AI algorithms based on digital and histopathological images of OSSN.
- Citation: Sinha S, Ramesh PV, Nishant P, Morya AK, Prasad R. Novel automated non-invasive detection of ocular surface squamous neoplasia using artificial intelligence. World J Methodol 2024; 14(2): 92267
- URL: https://www.wjgnet.com/2222-0682/full/v14/i2/92267.htm
- DOI: https://dx.doi.org/10.5662/wjm.v14.i2.92267
Artificial intelligence (AI) is the development of human or better than human abilities incorporated in computers, digital devices and machines to reduce quantum of error in the overall performance in a particular dimension. Since 1940, there has been rapid advancement in the utilization of AI in medical sciences, more so in the formulation of algorithms for effective diagnosis and management of various diseases. Ocular surface squamous neoplasia (OSSN) is among the common eye tumours responsible for considerable ocular morbidity causing sight-threatening complications if not diagnosed early and managed accordingly. Advanced invasive lesions may need orbital exenteration, which carries a risk of death. This article provides a comprehensive review of the deep and intricate insights regarding the role of AI in the non-invasive detection of OSSN, and presents the practical possibilities related to the future of AI in OSSN.
This review article was written after a literature search on EMBASE, Google Scholar, PubMed, Scopus, Web of Science and Cochrane library database. Only highly cited articles on OSSN and AI published between 1932 and 2022 in the English language were included. Reference Citation Analysis was also used to assess the articles on AI, relying upon the impact index per article to emphasize the latest published articles. The articles were reviewed by five independent researchers and the relevant literature compiled.
OSSN is an umbrella term used to include all primary epithelial dysplastic and carcinomatous lesions of the ocular surface (Table 1). It includes conjunctival or corneal intraepithelial neoplasia [conjunctival intraepithelial neoplasia (CIN): Wherein the neoplastic cells are confined to the epithelium and do not breach the basement membrane], as well as invasive squamous cell carcinoma (SCC) (invasive SCC: When the basement membrane is breached and neoplastic cells invade across it)[1-5].
| Term | Description | Structures included | First introduced |
| Epithelioma[1] | A generalized term encompassing neoplastic proliferation of the ocular surface epithelium, subsequently identified as squamous cell carcinoma of the conjunctiva and cornea | Conjunctiva and cornea | 1860 |
| Conjunctival Intraepithelial Neoplasia[2] | Abnormal neoplastic tissue involving the epithelium of the conjunctiva alone or the cornea as well | Conjunctiva and cornea | 1978 |
| Corneal Intraepithelial Neoplasia[2,3] | Disordered epithelial maturation (dysplasia) associated with abnormal growth of the corneal epithelium | Cornea | 1984 |
| Conjunctival and corneal invasive neoplasia[4] | Invasion of abnormal neoplastic tissue involving the epithelium of the conjunctiva or cornea | Conjunctiva or cornea | 1986 |
The overall incidence of OSSN has considerable geographical variation[6]. The incidence of invasive SCC varies from 0.02/100000 to 3.5/100000 population, and 75% of those affected are elderly, 75% are men and the lesion is limbal in 75% of the cases[7]. Low-income countries demonstrate the young, female demographic in OSSN prevalence, the higher-income demonstrate the older male demographic, with middle-income countries falling between the two with an intermediate age and no sex predilection[8]. In a recent study in India, the mean age at diagnosis was about 50 years[9]. In the elderly, they are the third most common type of oculo-orbital tumours, being exceeded in frequency only by melanoma and lymphoma[1]. OSSN poses a considerable economic burden, given the potentially high out-of-pocket costs for topical chemotherapy, lost work time for follow-up visits, the cost of the office visits, and even transportation costs to the visits[10].
The diagnosis of OSSN was initially entirely clinical. Patients with OSSN may be asymptomatic (30%) or present with non-specific symptoms like grittiness, redness, irritation and/or a mass in the eye which correlate poorly with the extent or depth of the lesion[5,11]. Multiple non-specific terms were used to describe the various presentations on observation of the ocular surface under direct illumination and magnification using a loupe or slit-lamp biomicroscope. The possibility of OSSN is suggested by a slightly elevated, usually well-demarcated fleshy lesion at the limbus with conjunctival part having a gelatinous, leukoplakic, or papillary appearance and feeding blood vessels usually in the interpalpebral area, a description which overlaps with several common inflammatory, dystrophic, or cicatricial disorders of the epibulbar region leading to wide differential diagnoses[12-14]. Similarly, corneal OSSN appears as an area of translucent clouding, which may not suggest a diagnosis of neoplasia at all[5]. In addition, due to varied differential diagnoses, it was not possible to rely entirely on history and clinical observation alone to exclude the possibility of malignancy in various ocular surface lesions, leading to the evolution of histopathologic methods for OSSN diagnosis[15].
Biopsy with classical staining methods was first done in 1932 where features of an OSSN lesion were studied and described to be of a papilloma[16]. However, McGavic and Stout studied the sections again, and identified it as corresponding to CIN. They concluded that this was the first report of histopathologically-proven Bowenoid epithelium occurring in the eye[17]. Ever since, histopathology remains the gold standard for confirmatory diagnosis of OSSN, with the use of staining of the specimen with vital dyes such as rose bengal, toluidine blue (0.05% or 1%) or methylene blue (1%) being a recent additional improvement[15,18-22]. The histopathological features were found to help in prognostication of OSSN and identify invasion of the basement membrane.
With the identification of viral associations of OSSN, efforts were directed to identify various viral etiological factors in OSSN which may coexist especially in immunodeficient patients[23]. Thus, Southern blot and in-situ hybridization (ISH) were then introduced as methods to detect human papillomavirus (HPV) infection in histopathological sections of ocular surface tumours in the 1980s[24,25]. It is of considerable interest, that it was found that ISH can distinguish reactive atypia and true dysplasia in conjunctival tumours[26]. However, RNA in formalin-fixed tissue is labile and likely to affect the sensitivity of ISH[27]. Only a weak association was found between HPV infection and OSSN[28].
Immunostaining for markers for proliferative potency and cell differentiation such as p16 and nuclear antigen Ki-67, mutant tumor suppressor gene p53, and argyrophilic nucleolar organizer regions was shown to be useful in detecting and prognosticating malignancy of conjunctival lesions including OSSN[29]. Preferential oncogene p63 expression has been demonstrated in anaplastic conjunctival epithelial cells with this technique[30]. However, these immunohistochemical approaches are prone to false-positive results as suggested recently[26].
Optical biopsy using a novel autofluorescence multispectral imaging technique has been recently described which produces false colour images from histopathological sections of ocular surface samples to enable identification of OSSN based on spectral signatures[31,32]. This is a promising technique for the identification of OSSN, which has a potential for automation. However, this too, cannot overcome the limitations of the excisional biopsy itself.
Excision biopsy is limited by the inability to determine orientation of the samples and subjectivity of the interpretation of histopathological findings. Lesions not included in the excised tissue may be missed as poorly demarcated diffuse lesions can be difficult to excise completely[27]. Moreover, with recurrent OSSN, repeated excisional biopsies may lead to conjunctival scarring and limbal stem cell deficiency[33]. The presence of negative histopathological margins is no guarantee for preventing the recurrence of OSSN[34]. This led to the exploration of non-invasive methods in the diagnosis of OSSN.
Direct dye staining of the lesion with vital dyes enables identification of OSSN in vivo, in very small lesions and in those coexisting with atopy and pterygium. However, dyes like lissamine green and rose bengal used to delineate the extent of OSSN lesions are nonspecific and stain many other ocular surface conditions[27]. Toluidine blue and methylene blue staining have high sensitivity but low to moderate specificity in diagnosing OSSN[20-22]. Thus, they are good as scree
Exfoliative cytology of conjunctival surface tumours had been popularized using platinum spatula, brush, and cotton-wool tipped swabs since 1954[35,36]. Shortcomings of exfoliative cytology included patient discomfort, inability to localize the pathology or study the degree of tumor invasion and altered cell-to-cell relationships causing inability to pinpoint the diagnosis of OSSN and not enabling differentiation between invasive and non-invasive disease[2]. Similar problems were encountered with aspiration cytology, in which cells were drawn from the conjunctival surface using a fluid-filled syringe[15]. It was in 1994 that impression cytology was employed to describe the diagnostic cytological findings and cell-to-cell relationships in cases of conjunctival tumours. Detailed cytomorphology of OSSN was then described in 2001[37]. Superficial epithelial cells can be collected by cellulose acetate paper, biopore membrane or millipore cellulose acetate filter paper to which the cells adhere and are lifted off the ocular surface. Impression cytology satisfactorily preserves the cytologic features, while overcoming the limitations of overlapping cells in exfoliative cytology specimens[38-40]. Cytology has an overall sensitivity of 72.4% and a specificity of 74.3%, and a considerable proportion of cytology specimens may have to be excluded from analysis due to inadequate cellularity[41]. Impression cytology has been found to be positive in only 77% to 80% of cases as it can assess only superficial cells and is unable to sample deep lesions or invasive disease[40]. It is not possible to differentiate intraepithelial lesions from invasive SCC given the superficial sampling of cells in this technique[37,42,43]. Moreover, it requires considerably skilled professionals to interpret results. A fair negative predictive accuracy indicates that impression cytology is a valuable screening technique, but not a gold standard[42]. Staining of cytology specimens with vital dyes is another improvement, however, it could not overcome the low specificity that makes it only a good screening tool[18-22].
In 2002, Char et al[44] first reported ultrasound biomicroscopy (UBM) for studying SCC of the ocular surface with intraocular extension. On UBM, the surface of the OSSN is hyperechoic while the interior is hypoechoic. It was found to be the most useful tool to assess and document intraocular extension of OSSN, wherein blunting of the anterior chamber angle and uveal thickening are evident findings which correlate well with histopathological observations[45]. UBM requires a water bath in the reclined position and a technician familiar with its use to obtain the best images. Moreover, for studying intraocular invasion of OSSN, it requires two probes: A 50 MHz probe for images that have better resolution, and a 20 MHz UBM to provide deeper and wider field of view. However, definition of the posterior margins of the tumor is limited by maximum penetration of both 20 MHz and 50 MHz UBM[45]. Images have lower resolution as compared to the newer modalities[27].
Duchateau et al[46] first applied in vivo confocal microscopy (IVCM) to assess CIN in 2005. Subsequently, various authors described different features of OSSN on IVCM, some of which could provide microscopic evidence of corneal involvement[47]. IVCM can identify cellular anisocytosis, anisonucleosis, altered nuclear:cytoplasmic ratio, and nests of isolated keratinized mitotic cells extending beyond the basement membrane (invasion)[48]. Several distinctive features such as large nucleoli with starry night appearance and the absence of sub-basal corneal nerves make IVCM diagnostic for OSSN, having complete agreement with scraping cytology and histology[49,50]. However, the instrument is limited in availability, and the quality of imaging relies on the illumination of the object and reflected light. Ocular surface tumours have poor optical properties given the keratinization, especially the leukoplakic types. And several features of benign and malignant lesions on IVCM overlap, making it difficult to guide treatment decisions[15]. Confocal microscopy requires a skilled technician to perform the test[27]. Moreover, it is neither possible to determine the presence of microinvasion by this method, nor is it possible to reliably differentiate benign from OSSN lesions due to overlap of IVCM features in various ocular pathologies[46]. Additionally, it is difficult to obtain IVCM images and biopsy specimens from the exact same site where the tissue is being examined[51]. Another limitation is that it can provide only en-face images in contrast to cross-sectional images obtained in tissue histology[50]. Moreover, because it provides images at a cellular level, IVCM is unable to provide a comprehensive scan of the entire ocular surface[27].
Anterior segment optical coherence tomography shows epithelial thickening and increased reflectivity of epithelium with an obvious delineation of OSSN-affected tissue, which correlates well with the gold standard histopathologic examination in differentiating OSSN from other corneal and conjunctival pathologies to guide appropriate management and monitor treatment response[15,52,53]. Microscopic detail to the extent of a 2 µm resolution was achieved with an ultrahigh-resolution optical coherence tomography (UHR-OCT) device in 2011 and its role in monitoring the disease, detection of recurrence and guiding the withdrawal of treatment has been well documented[33]. UHR-OCT is useful when clinical differential diagnosis of ocular surface lesions are broad, not only in the case of small limbal OSSN which are frequently misdiagnosed as pterygia, but also in the case of corneal OSSN which can be clinically confused with Salzmann nodular degeneration, vascularized corneal scars, melanomas, sequelae of limbal stem cell deficiency, herpes simplex viral keratopathy, atypical peripheral corneal infiltrates (suspicious for vernal keratoconjunctivitis) and other complex ocular surface pathology which may even coexist with OSSN[54]. Optical signs indicating complex, subtle, atypical or residual OSSN are revealed by UHR-OCT which shows tremendous promise for the non-invasive diagnosis of OSSN[33,54]. The UHR-OCT is, however, a custom built machine, and is not commonly available[27].
The use of commercially available, HR, spectral-domain OCT in 2015 was a giant leap for the non-invasive diagnosis of OSSN. OCT can be used to measure epithelial thickness, which differentiates OSSN from pterygium with an 87%-100% sensitivity and 75%-100% specificity[41,52]. OCT angiography imaging on newer OCT devices allows for visualization and quantification of vessel structure and density within, under, and surrounding OSSN[55]. Mean vessel area density in the subepithelium adjacent to the OSSN increases with treatment, then decreases significantly between mid-treatment and resolution[56]. This can be useful in evaluating the effect of medical therapy on OSSN resolution.
Modern OCT modalities are not without limitations as they are less useful in evaluating pigmented lesions[52]. There are no consistent criteria for distinguishing CIS from invasive SCC on OCT. The presence of feeder vessels, intrinsic vascularity and a nodular lesion raise suspicion of invasive SCC, but there are no objective or quantitative criteria regarding their presence on OCT which may suggest invasion. In addition, the depth of invasion of OSSN cannot be reliably assessed due to poor penetration of the OCT[57]. OCT angiographic studies are limited by artifacts created by patient movements. Thick tumors and leukoplakia preclude detailed visualization of blood vessels especially in the subepithelial tissue. The relationships between baseline and change in vessel characteristics and tumor grade and stage are poorly characterized. In addition, there are difficulties in image processing, including manual segmentation of the tumors, projection artifacts, and vascular density calculations[56].
The multiple modalities and imaging data from invasive and non-invasive methods of detection of OSSN are complex to interpret and correlate with patient characteristics, history and clinical examination findings, response to treatment, follow-up and final outcomes. These results can be observed by humans as well as AI algorithms, which raises the possibility of automating the diagnosis of OSSN.
AI has emerged as a powerful tool in various fields, and its potential in OSSN diagnosis is particularly promising. With its ability to process vast amounts of data and identify complex patterns, AI has the potential to revolutionize non-invasive objective diagnosis of OSSN based on automation, and improve patient outcomes. This is possible using various component systems working together to function like human intelligence, while avoiding the factors of subjectivity, fatigue and bias.
Neural networks enable computers to perform complex tasks once thought to be the exclusive domain of human intelligence. Two prominent architectures of artificial neural networks (ANN) and convolutional neural networks (CNN) have made significant contributions to the field of machine learning, enabling breakthroughs in the diagnosis of breast cancer, skin cancer, prostate cancer, lung cancer, etc[58].
ANN are inspired by the structure and function of biological neural networks in the human brain. ANN consist of interconnected nodes, called artificial neurons or perceptrons, organized into layers, the three main types of layers being the input layer, hidden layers, and output layer. Each neuron receives inputs, applies an activation function, and passes the result to the perceptrons in the next layer. Through a process called backpropagation, neural networks can learn from data and adjust the weights of their connections to improve their performance.
CNN, on the other hand, are specifically designed for processing grid-like data such as images, and excel in tasks that require extracting meaningful features from input data. They use a specialized type of convolutional neuron to perform convolution operations on the input data. CNN typically consist of multiple layers, including convolutional layers, pooling layers, and fully connected layers. The convolutional layers extract local patterns or features from the input data, while the pooling layers downsample the feature maps to reduce the spatial dimensions. The fully connected layers at the end of the network perform classification or regression tasks based on the extracted features.
The strength of ANN lies in its ability to solve a wide range of problems, including regression, classification, and sequence generation enabling tasks like image recognition. On the other hand, CNN have demonstrated exceptional performance in computer vision tasks, such as object detection, image segmentation, and image classification due to the ability to capture local patterns and spatial hierarchies. These have already been successful in classifying cancerous lesions in organs other than the eyes, thus building promise for their use in OSSN[59].
Deep learning focuses on training ANN with multiple layers, known as deep neural networks, which mimic the complex interconnections and hierarchical representations found in the human brain. Deep learning algorithms learn directly from raw data by automatically extracting features and patterns through multiple layers of computation. These layers allow for increasingly abstract representations of the input data, enabling the network to learn hierarchical features and make more accurate predictions or decisions. One of the key advantages is its ability to handle large-scale, high-dimensional data. As of today, various types of medical images, such as those of radiological investigations have been used to develop deep learning models for cancers of the lung, rectum, pancreas, stomach, prostate, brain, breast, etc[60].
Data augmentation is a technique widely used in machine learning and deep learning to increase the size and diversity of a training dataset by creating new samples through various transformations. It involves applying random modifications to the existing data, such as rotations, translations, flips, zooms, or changes in brightness and contrast. Its purpose is to improve the generalization and robustness of machine learning models. It is particularly useful in scenarios where the available labelled data is limited or imbalanced. By providing additional variations of the training data, it helps the model learn more invariant and discriminative features, reducing overfitting and enhancing the model's ability to handle unseen data.
Commonly used in computer vision tasks, such as image classification or object detection, data augmentation can also be applied to other types of data. By artificially increasing the size and diversity of the training dataset, this serves as a regularization technique enabling improved performance and better utilization of available data resources.
Pre-trained networks are ANN that have been trained on large-scale datasets for specific tasks, such as image classification or natural language processing. The concept behind these networks is to leverage the knowledge and features learned from one task and transfer them to another related task. By initializing the network with pre-trained weights, the model already possesses some understanding of the underlying patterns in the data, saving significant training time and resources. Hence, they can achieve state-of-the-art performance on various tasks, even with limited labelled data. They also facilitate faster experimentation and prototyping, as researchers can focus on fine-tuning the model for specific tasks rather than starting from scratch.
Common pre-trained network models like very deep neural networks, deep residual networks, and bidirectional encoder representation from transformers have been pre-trained on large-scale datasets like ImageNet, capturing general features and patterns. Researchers and practitioners can then adapt these pre-trained models by adding or modifying layers to suit their specific task or domain.
Feature extractors are algorithms or components within machine learning systems responsible for capturing and representing meaningful patterns or features from raw data. This is important when dealing with high-dimensional or complex data, such as images, audio, or text. They can reduce the dimensionality of the data, remove noise or irrelevant information, and highlight discriminative characteristics essential for the learning process. Feature extractors can be handcrafted, or they can be learned automatically through deep learning techniques.
Transfer learning is a machine learning technique that leverages knowledge and models learned from one task or domain to improve performance on a different but related task or domain. Pre-trained models trained on large datasets for a specific task can be adapted to new tasks or datasets. Transfer learning can significantly reduce the amount of labelled data required for training, making it useful when labelled data is scarce or expensive to obtain. Overall, it enables researchers and practitioners to transfer knowledge and representations across tasks, leading to improved performance, faster training, and better utilization of available resources. As of today, transfer learning is being widely used in interpretation of retinal scans obtained by modern imaging techniques like optical coherence tomography angiography[61].
AI algorithms are increasingly being utilized for both qualitative and quantitative analysis in various domains. In qualitative analysis, AI algorithms can be used to analyze unstructured data, such as text or images, and extract relevant information. Computer vision algorithms can interpret images and videos, enabling object recognition, and image classification. On the other hand, AI algorithms play a crucial role in quantitative analysis by processing structured data and performing statistical analyses. These can identify patterns, correlations, and trends as well as perform predictive modelling, regression analysis, and clustering, providing valuable insights into complex datasets. AI algorithms also enable the integration of qualitative and quantitative analyses by combining different types of data.
One of the key challenges in OSSN diagnosis is the accurate interpretation of medical images, such as anterior segment photographs, OCT images, computed tomography and magnetic resonance imaging scans, and those obtained by other in vivo techniques. AI algorithms can analyze these non-invasively acquired images with incredible speed and precision, assisting radiologists, pathologists and ophthalmic surgeons in detecting subtle abnormalities that may indicate the presence and extent of neoplasia. AI can also help in distinguishing benign lesions from malignant tumours, reducing the need for unnecessary biopsies and surgeries.
AI can aid in early detection of OSSN, leading to better treatment outcomes. By analyzing patient data, including genetics, medical history and lifestyle factors, the AI algorithms can identify individuals at a higher risk of developing OSSN and determine the interacting risk factors in a patient with a suspicious lesion. This enables proactive screening and early intervention, when the disease is more treatable and potentially curable.
Another area of great promise is in the development of personalized treatment plans. OSSN is a complex disease with significant inter-patient variability. AI algorithms can integrate patient-specific data and clinical knowledge to predict treatment responses and recommend tailored therapies. This can optimize treatment decisions between various possible approaches, improve rates of treatment success, and minimize unnecessary side effects.
Furthermore, AI-powered tools can enhance the efficiency and accuracy of molecular profiling crucial for precision medicine. AI algorithms can analyze large genomic datasets, identify genetic mutations or biomarkers associated with specific cancers, and suggest targeted therapies or clinical trials. This will enable ocular oncologists to make more informed decisions about the most effective treatment options for individual patients.
AI can also assist in monitoring treatment responses. By continuously analysing images, laboratory results, and real-time physiological data, AI algorithms can detect early signs of treatment failure or disease progression. This facilitates timely adjustments to treatment plans, ensuring optimal outcomes.
Automated screening aims to detect and identify suspicious lesions on the ocular surface[62]. The integration of AI in automated screening for OSSN brings several advantages (Table 2).
| Advantages of automated screening by AI for OSSN |
| It enables the screening process to be more efficient and objective, reducing the reliance on subjective human interpretation |
| AI algorithms can analyze large volumes of images rapidly and consistently, aiding in the early identification of suspicious lesions that may otherwise be overlooked |
| Automated screening can enhance access to care, particularly in areas where specialized ophthalmic expertise may be limited, through telemedicine and remote monitoring |
| By providing a preliminary assessment of OSSN lesions, AI technology can support primary care providers and community healthcare workers in triaging patients and referring those in need of further evaluation to specialized centers |
| AI-driven automated screening holds promise in improving the early detection and management of OSSN, ultimately leading to better patient outcomes |
AI can highlight areas of potential abnormality in the ocular surface for further evaluation. By leveraging machine learning techniques, AI models can learn from a vast dataset of annotated images to accurately differentiate OSSN lesions from normal ocular tissue[63].
AI models can analyze clinical and molecular data to predict outcomes such as treatment response, disease progression, and recurrence. By considering patient-specific factors and integrating multimodal data, AI can generate personalized predictions that enable clinicians to optimize treatment strategies, adjust follow-up schedules, and tailor interventions based on the individual characteristics of each patient, ultimately improving the overall management and prognosis of OSSN[64].
By analyzing genetic data from tissue samples, AI algorithms can identify patterns and correlations that may be indicative of aggressive disease or potential treatment targets[65]. This molecular profiling combined with predictive modelling could enable a deeper understanding of the underlying mechanisms of OSSN and facilitate the development of targeted therapies.
Furthermore, AI predictions may assist in stratifying OSSN subtypes, which may have different clinical behaviours and responses to treatment. This stratification can allow for more precise and personalized therapeutic approaches, potentially reducing unnecessary interventions and improving patient outcomes[51].
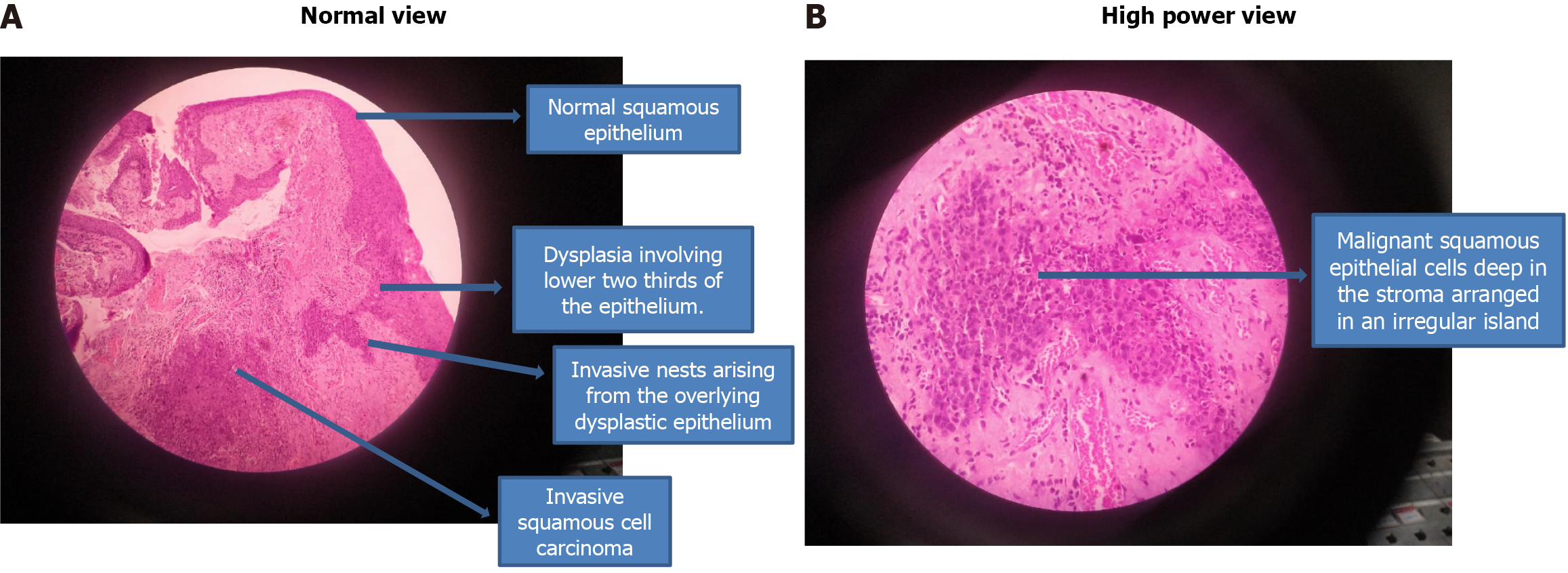
Deep learning models based on ANN with multiple layers, can analyze large amounts of data, including images, clinical features, and molecular information, to make accurate and efficient diagnostic predictions for OSSN. By training on annotated datasets of OSSN cases, deep learning models can learn complex patterns and features that are indicative of the disease, enabling them to differentiate between normal ocular tissue and OSSN lesions (Figure 1). This holds great potential for improving diagnostic accuracy, reducing interobserver variability, and assisting healthcare professionals in making informed decisions[66].
In the case of OSSN, deep learning models can analyze images of the ocular surface to detect irregularities in tissue morphology, abnormal vascular patterns, and other features that may be challenging for human observers to discern. By leveraging CNN which are particularly adept at image analysis, deep learning models can automatically extract relevant features from OSSN images and provide objective and consistent diagnostic assessments[48].
AI algorithms can be trained to accurately classify OSSN lesions into different subtypes based on their characteristics, such as size, location, and extent of involvement. By analyzing clinical and imaging data, including photographs, OCT scans, and histopathological features, AI models can categorize OSSN lesions into distinct subtypes, such as intraepithelial neoplasia, invasive SCC, or carcinoma in situ[67].
Furthermore, AI-based automated staging of OSSN lesions can aid in determining the extent and severity of the disease. By analyzing imaging data and clinical parameters, AI models can predict the stage of OSSN, which refers to the depth of invasion and potential spread of the neoplastic cells. Thus, it helps identify cases that may require more aggressive interventions, such as surgical excision or adjuvant therapy. By accurately predicting the stage of OSSN, AI models can assist clinicians in developing personalized treatment plans[33].
AI-based evaluation of severity in OSSN offers several benefits (Table 3). AI algorithms can analyze various clinical and imaging data to assess the severity of OSSN lesions accurately. By utilizing machine learning techniques, AI models can learn from annotated datasets and develop algorithms that consider factors such as lesion size, location, extent of invasion, and involvement of adjacent structures. This comprehensive evaluation of severity aids in determining the appropriate treatment approach, including the need for surgical intervention, adjuvant therapy, or conservative management[68].
| Advantages of AI-based evaluation of severity in OSSN |
| It provides an objective and standardized assessment, reducing interobserver variability that may be present in traditional evaluation methods AI algorithms can analyze large amounts of data rapidly and consistently, ensuring accurate and reproducible severity evaluation |
| Additionally, AI can incorporate multi-modal data, including imaging findings from techniques such as OCT, confocal microscopy, or histopathological characteristics |
| This integration of diverse data sources enhances the accuracy and reliability of severity evaluation, enabling clinicians to make informed decisions regarding treatment planning and prognostication |
| Overall, AI-driven evaluation of severity in OSSN holds promise in improving patient outcomes by facilitating appropriate and tailored management strategies based on the individual characteristics of each case |
Patients who undergo diagnostic evaluation using non-invasive AI-based tools in conjunction with expert clinical decision-making by ocular oncologists are likely to experience more rapid, reliable, and robust interpretation of their clinical condition, assurance of a better clinical response to treatment modalities selected and a more complete explanation of the prognosis and risk of recurrence, leading to better patient experience. However, enforcing AI-based treatment decisions may undermine the participation of the patient in the decision-making process, leading to poor acceptance of these treatment modalities and follow-up regimens, with overall less satisfaction and potential drop-out or loss to follow-up. For example, an AI algorithm might predict that a certain OSSN may respond better to Mitomycin C drops compared to interferon alpha 2B drops, but the ocular surface irritation caused by mitomycin drops may lead to poor compliance to this therapy leading to poor outcome and less patient satisfaction[62].
Despite the numerous advantages of AI in OSSN, certain limitations and challenges need to be acknowledged. Firstly, the availability and quality of data are crucial for training AI models. In the case of OSSN, obtaining large, diverse, and well-annotated datasets can be challenging, as the condition is relatively rare. Limited data availability may affect the performance and generalizability of AI algorithms. Additionally, the development and validation of robust AI models requires substantial computational resources, technical expertise, and rigorous testing. Ensuring the reliability, accuracy, and safety of AI algorithms is of utmost importance to avoid potential errors or biases. Multicentric image acquisition with centralized image processing and expert interpretation may be useful to train the data sets to develop the AI models. Identification of key centres and international collaborations to include diverse ethnic groups should be the first step towards overcoming these challenges.
Ethical considerations, such as privacy concerns and the responsible use of patient data, must be addressed when implementing AI in OSSN. The concerns encompass breach of patient identity, hacking of ocular signatures such as iris patterns, and even misuse of genetic information. To prevent attacks such as these, data must be processed through the use of secure and encrypted channels, and the results transferred in an anonymized manner to protect patient identity and privacy. Research should explore how cloud computing and blockchain technology can be made useful for addressing concerns of data security in this regard[69].
Furthermore, the integration of AI technology into clinical practice requires proper validation, training, education, and acceptance from healthcare professionals to effectively leverage its benefits through collaborative efforts among clinicians, researchers, and AI experts. Research and publications of increasingly higher levels of evidence will help to overcome the concerns about shortcomings of AI models and improve their reliability and acceptance.
Computer-assisted image analysis is the first step towards application of AI in OSSN. By leveraging AI algorithms, computer systems can analyze images of the ocular surface to aid in the detection, characterization, and monitoring of OSSN lesions in large populations[70,71]. This technology enables enhanced visualization and analysis of OSSN lesions, facilitating early detection, precise diagnosis, and improved treatment planning[34].
One of the key advantages of computer-assisted image analysis is its ability to detect subtle and early changes in OSSN lesions that may not be apparent to the human eye[72]. Furthermore, computer-assisted image analysis provides a standardized and reproducible approach, reducing interobserver variability and enhancing the consistency and reliability of OSSN assessment[73].
Several deep learning frameworks have emerged that can facilitate the development and deployment of deep neural networks in OSSN research and clinical practice. For example, frameworks such as TensorFlow, PyTorch, and Keras offer a wide range of pre-built modules, functions, and tools that streamline the implementation of deep learning models, which provide a user-friendly interface and enable efficient model training and evaluation, allowing researchers and clinicians to explore new algorithms, validate research findings, and develop innovative solutions to improve the diagnosis and management of OSSN[74-76].
Validation of AI against expert panels is a crucial step in assessing the performance and reliability of AI algorithms in OSSN. The expert panel could consist of ophthalmologists, pathologists, and other specialists who have extensive knowledge and expertise in diagnosing and managing OSSN. By comparing the AI-generated assessments with the consensus decisions of the expert panel, the accuracy, sensitivity, specificity, positive predictive value and negative predictive value of the AI algorithm can be evaluated[5,77]. The validation process also highlights areas where the algorithm may outperform or fall short compared to human experts. This iterative process of validation and refinement is essential for optimizing AI algorithms, improving their accuracy, and gaining confidence in their reliability for clinical applications in OSSN[78].
Need for shift in protocols: There is a growing need for a shift in protocols regarding the incorporation of AI algorithms in OSSN management. Traditional protocols often rely on manual evaluation and subjective interpretation by healthcare professionals, which can be time-consuming and prone to interobserver variability[79]. The integration of AI can streamline and enhance these protocols by providing objective, consistent, and efficient analysis of OSSN lesions. By leveraging AI's capabilities in image analysis, pattern recognition, and predictive modelling, protocols can be modified to include automated screening, diagnosis, classification, severity evaluation, and treatment prediction[14].
Furthermore, the implementation of AI in OSSN protocols can lead to improved efficiency and resource allocation due to faster screening, reducing patient wait times and optimizing clinical workflow. Moreover, identifying high-risk cases that require more immediate attention allows for timely interventions[80]. By integrating AI into protocols, healthcare providers can allocate their expertise and resources more effectively, focusing on complex cases and providing personalized care while AI handles routine tasks. Thus, it has the potential to enhance the overall efficiency, quality, and accessibility of OSSN management.
Phase of testing compliance of computer based image analysis: This phase involves ensuring that the AI algorithms and systems meet regulatory standards and compliance requirements. This phase focuses on evaluating their safety, reliability, accuracy, precision, robustness, and generalizability[81]. It also involves assessing the security and privacy aspects. Compliance testing aims to mitigate risks, validate the effectiveness of the technology, and gain regulatory approvals or certifications necessary for its use in clinical settings[82].
During compliance testing, the AI algorithms and systems are subjected to various validation and verification processes involving comparison of AI-generated results with ground truth data, conducting performance evaluations, and assessing the system's ability to handle different scenarios and variations[83]. Compliance testing also includes conducting comprehensive risk assessments to identify and address potential vulnerabilities or biases in the AI algorithms. Additionally, the systems' compliance with privacy regulations, such as data anonymization and encryption, is evaluated to ensure the protection of patient confidentiality.
Phase of initial acceptance: During this phase, healthcare professionals and institutions begin to integrate AI algorithms and systems into their workflows and protocols. The initial acceptance phase focuses on user acceptance, training, and education to ensure the successful implementation and utilization of the AI technology. Healthcare providers need to be familiarized with the capabilities, limitations, and benefits of the AI systems, as well as their integration into existing clinical processes[84]. Training may include workshops, demonstrations, and hands-on sessions to educate users. Additionally, clear guidelines and protocols are established to outline the roles and responsibilities of healthcare professionals when using AI systems, ensuring proper integration into clinical practice. Continuous support, feedback mechanisms, and collaborative efforts between AI developers and healthcare providers are crucial during this phase to address any challenges or concerns and facilitate a smooth transition. This phase marks the beginning of the widespread adoption of computer-based image analysis in OSSN, bringing the benefits of AI technology to the forefront of clinical decision-making and patient care[85].
Phase of full utilization: The phase of full utilization represents the culmination of the integration of computer-based image analysis in OSSN management. During this, AI technology will be fully embraced and incorporated into routine clinical practice for OSSN. Its utilization will become an integral part of the standard workflow, diagnosis and management, reflecting the optimization of processes and outcomes through the effective utilization of AI technology[86].
In the phase of full utilization, healthcare providers would have gained extensive experience and expertise in working with the AI systems. They would have developed a deep understanding of the technology's capabilities, limitations, and potential applications. Refinements would be based on user feedback, continuous learning, and advancements in the field through continuous quality improvement initiatives. Full utilization of computer-based image analysis will enable healthcare providers to leverage the benefits of AI for OSSN management, such as improved accuracy, efficiency, and standardization, leading to enhanced patient care, better treatment outcomes, and more effective resource allocation (Table 4)[87].
| Advantages of integrated AI in OSSN |
| AI algorithms would analyze large volumes of data with speed and accuracy, surpassing human capabilities in terms of processing efficiency |
| This capability would allow for rapid and efficient screening, diagnosis, and evaluation of OSSN lesions, saving valuable time and resources for healthcare professionals |
| AI models would progressively learn from vast datasets, enabling them to identify increasingly complex patterns and subtle features that may be challenging for human observers to detect |
| This ability would offer enhanced diagnostic accuracy and aid in early detection, potentially improving patient outcomes and prognosis |
| AI would provide a standardized and objective assessment, reducing interobserver variability and ensuring consistent and reliable evaluations of severity, classification, and staging |
| By leveraging AI technology, clinicians would benefit from enhanced decision support, optimized treatment planning, and personalized management strategies for OSSN patients |
The future of deep learning-based image analysis in OSSN holds immense potential for further advancements and transformative impacts. With the availability of larger and more comprehensive datasets, deep learning models can be trained to extract even more nuanced features and patterns from OSSN images, leading to improved diagnostic accuracy and precision.
Additionally, the integration of multimodal data, such as combining imaging information with molecular profiling or clinical data, can enable more precise risk stratification, treatment prediction, and personalized management approaches. Furthermore, the utilization of real-time data and the incorporation of dynamic imaging techniques, such as video-based analysis or sequential image analysis, can provide valuable insights into the temporal changes and progression of OSSN, supporting more proactive and timely interventions[88].
AI algorithms can be leveraged for treatment planning, monitoring treatment response, and predicting long-term outcomes. By integrating deep learning with clinical and genetic data, personalized treatment plans can be developed, optimizing therapeutic strategies and improving patient outcomes. Moreover, the development of user-friendly interfaces, integration with electronic health records, and advancements in telemedicine technologies can facilitate the widespread adoption and utilization of deep learning-based image analysis in OSSN, allowing for seamless integration into routine clinical practice and improving access to specialized care.
Overall, the future of deep learning-based image analysis in OSSN holds great promise in revolutionizing the field, enabling more accurate diagnoses, personalized treatment approaches, and improved patient care.
| 1. | Basti S, Macsai MS. Ocular surface squamous neoplasia: a review. Cornea. 2003;22:687-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 152] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 2. | Hamam R, Bhat P, Foster CS. Conjunctival/corneal intraepithelial neoplasia. Int Ophthalmol Clin. 2009;49:63-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 3. | Waring GO 3rd, Roth AM, Ekins MB. Clinical and pathologic description of 17 cases of corneal intraepithelial neoplasia. Am J Ophthalmol. 1984;97:547-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 85] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 4. | Erie JC, Campbell RJ, Liesegang TJ. Conjunctival and corneal intraepithelial and invasive neoplasia. Ophthalmology. 1986;93:176-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 213] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 5. | Lee GA, Hirst LW. Retrospective study of ocular surface squamous neoplasia. Aust N Z J Ophthalmol. 1997;25:269-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Gurnani B, Kaur K. Ocular Surface Squamous Neoplasia. 2023 Jul 31. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. [PubMed] |
| 7. | Honavar SG, Manjandavida FP. Tumors of the ocular surface: A review. Indian J Ophthalmol. 2015;63:187-203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Hӧllhumer R, Michelow P, Williams S. Demographics, clinical presentation and risk factors of ocular surface squamous neoplasia at a tertiary hospital, South Africa. Eye (Lond). 2023;37:3602-3608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 9. | Kaliki S, Vempuluru VS, Ghose N, Gunda S, Vithalani NM, Sultana S, Ganguly A, Bejjanki KM, Jakati S, Mishra DK. Ocular surface squamous neoplasia in India: a study of 438 patients. Int Ophthalmol. 2022;42:1915-1926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 10. | Saffra NA, Emborgo TS, Iacob CE, Kirsch DS. Cost-effective treatment of ocular surface squamous neoplasia for an undocumented and uninsured New York City patient: a case report. J Med Case Rep. 2020;14:174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 11. | Winter FC, Kleh TR. Precancerous epithelioma of the limbus. Arch Ophthalmol. 1960;64:208-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 12. | Irvine AR Jr. Epibulbar squamous cell carcinoma and related lesions. Int Ophthalmol Clin. 1972;12:71-83. [PubMed] |
| 13. | Brown HH, Glasgow BJ, Holland GN, Foos RY. Keratinizing corneal intraepithelial neoplasia. Cornea. 1989;8:220-224. [PubMed] |
| 14. | Sayed-Ahmed IO, Palioura S, Galor A, Karp CL. Diagnosis and Medical Management of Ocular Surface Squamous Neoplasia. Expert Rev Ophthalmol. 2017;12:11-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 77] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 15. | Nanji AA, Mercado C, Galor A, Dubovy S, Karp CL. Updates in Ocular Surface Tumor Diagnostics. Int Ophthalmol Clin. 2017;57:47-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (1)] |
| 16. | Weymann MF. Diffuse Papilloma of the Limbus. Am J Ophthalmol. 1932;15:310-312. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 17. | McGavic JS. Intraepithelial Epithelioma of the Cornea and Conjunctiva (Bowen’s Disease)*. Am J Ophthalmol. 1942;25: 167-176. [RCA] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Wilson FM 2nd. Rose bengal staining of epibulbar squamous neoplasms. Ophthalmic Surg. 1976;7:21-23. [PubMed] |
| 19. | Kaji Y, Hiraoka T, Oshika T. Vital staining of squamous cell carcinoma of the conjunctiva using toluidine blue. Acta Ophthalmol Scand. 2006;84:825-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Gichuhi S, Macharia E, Kabiru J, Zindamoyen AM, Rono H, Ollando E, Wanyonyi L, Wachira J, Munene R, Onyuma T, Jaoko WG, Sagoo MS, Weiss HA, Burton MJ. Toluidine Blue 0.05% Vital Staining for the Diagnosis of Ocular Surface Squamous Neoplasia in Kenya. JAMA Ophthalmol. 2015;133:1314-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Romero IL, Barros Jde N, Martins MC, Ballalai PL. The use of 1% toluidine blue eye drops in the diagnosis of ocular surface squamous neoplasia. Cornea. 2013;32:36-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Steffen J, Rice J, Lecuona K, Carrara H. Identification of ocular surface squamous neoplasia by in vivo staining with methylene blue. Br J Ophthalmol. 2014;98:13-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Simbiri KO, Murakami M, Feldman M, Steenhoff AP, Nkomazana O, Bisson G, Robertson ES. Multiple oncogenic viruses identified in Ocular surface squamous neoplasia in HIV-1 patients. Infect Agent Cancer. 2010;5:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Lass JH, Jenson AB, Papale JJ, Albert DM. Papillomavirus in human conjunctival papillomas. Am J Ophthalmol. 1983;95:364-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 29] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 25. | Naghashfar Z, McDonnell PJ, McDonnell JM, Green WR, Shah KV. Genital tract papillomavirus type 6 in recurrent conjunctival papilloma. Arch Ophthalmol. 1986;104:1814-1815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 26. | Peterson C, Parikh RN, Ahmad MT, Campbell AA, Daoud Y, Mahoney N, Siadati S, Eberhart CG. Detection of Human Papillomavirus in Squamous Lesions of the Conjunctiva Using RNA and DNA In-Situ Hybridization. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 27. | Ong SS, Vora GK, Gupta PK. Anterior Segment Imaging in Ocular Surface Squamous Neoplasia. J Ophthalmol. 2016;2016:5435092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 28. | Shrestha T, Choi W, Kim GE, Yang JM, Yoon KC. Human papilloma virus identification in ocular surface squamous neoplasia by p16 immunohistochemistry and DNA chip test: A strobe-compliant article. Medicine (Baltimore). 2019;98:e13944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Aoki S, Kubo E, Nakamura S, Tsuzuki A, Tsuzuki S, Takahashi Y, Akagi Y. Possible prognostic markers in conjunctival dysplasia and squamous cell carcinoma. Jpn J Ophthalmol. 1998;42:256-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 30. | Auw-Haedrich C, Sundmacher R, Freudenberg N, Spelsberg H, Feltgen N, Maier P, Reinhard T. Expression of p63 in conjunctival intraepithelial neoplasia and squamous cell carcinoma. Graefes Arch Clin Exp Ophthalmol. 2006;244:96-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Habibalahi A, Bala C, Allende A, Anwer AG, Goldys EM. Novel automated non invasive detection of ocular surface squamous neoplasia using multispectral autofluorescence imaging. Ocul Surf. 2019;17:540-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 32. | Habibalahi A, Allende A, Michael J, Anwer AG, Campbell J, Mahbub SB, Bala C, Coroneo MT, Goldys EM. Pterygium and Ocular Surface Squamous Neoplasia: Optical Biopsy Using a Novel Autofluorescence Multispectral Imaging Technique. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 33. | Shousha MA, Karp CL, Canto AP, Hodson K, Oellers P, Kao AA, Bielory B, Matthews J, Dubovy SR, Perez VL, Wang J. Diagnosis of ocular surface lesions using ultra-high-resolution optical coherence tomography. Ophthalmology. 2013;120:883-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 34. | Tabin G, Levin S, Snibson G, Loughnan M, Taylor H. Late recurrences and the necessity for long-term follow-up in corneal and conjunctival intraepithelial neoplasia. Ophthalmology. 1997;104:485-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 164] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 35. | Larmande A, Timsit E. Importance of cytodiagnosis in ophthalmology: preliminary report of 8 cases of tumors of the sclero-corneal limbus. Bull Soc Ophtalmol Fr. 1954;5:415-419. [PubMed] |
| 36. | Pe’er J. Ocular Surface Squamous Neoplasia. Ophthalmol Clin N Am. 2005;18:1-13. [RCA] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 37. | Nolan GR, Hirst LW, Bancroft BJ. The cytomorphology of ocular surface squamous neoplasia by using impression cytology. Cancer. 2001;93:60-67. [PubMed] |
| 38. | Gelender H, Forster RK. Papanicolaou cytology in the diagnosis and management of external ocular tumors. Arch Ophthalmol. 1980;98:909-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 39. | Nolan GR, Hirst LW, Wright RG, Bancroft BJ. Application of impression cytology to the diagnosis of conjunctival neoplasms. Diagn Cytopathol. 1994;11:246-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 40. | Tole DM, McKelvie PA, Daniell M. Reliability of impression cytology for the diagnosis of ocular surface squamous neoplasia employing the Biopore membrane. Br J Ophthalmol. 2001;85:154-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 64] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 41. | Hӧllhumer R, Michelow P, Williams S. Comparison of non-invasive diagnostic modalities for ocular surface squamous neoplasia at a tertiary hospital, South Africa. Eye (Lond). 2024;38:1118-1124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 42. | Tananuvat N, Lertprasertsuk N, Mahanupap P, Noppanakeepong P. Role of impression cytology in diagnosis of ocular surface neoplasia. Cornea. 2008;27:269-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 43. | Barros JN, Lowen MS, Ballalai PL, Mascaro VL, Gomes JA, Martins MC. Predictive index to differentiate invasive squamous cell carcinoma from preinvasive ocular surface lesions by impression cytology. Br J Ophthalmol. 2009;93:209-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 44. | Char DH, Kundert G, Bove R, Crawford JB. 20 MHz high frequency ultrasound assessment of scleral and intraocular conjunctival squamous cell carcinoma. Br J Ophthalmol. 2002;86:632-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 45. | Finger PT, Tran HV, Turbin RE, Perry HD, Abramson DH, Chin K, Della Rocca R, Ritch R. High-frequency ultrasonographic evaluation of conjunctival intraepithelial neoplasia and squamous cell carcinoma. Arch Ophthalmol. 2003;121:168-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 46. | Duchateau N, Hugol D, D'Hermies F, Meyer A, Labbé A, Dupas B, Iordanidou V, Renard G, Baudouin C. [Contribution of in vivo confocal microscopy to limbal tumor evaluation]. J Fr Ophtalmol. 2005;28:810-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 47. | Balestrazzi A, Martone G, Pichierri P, Tosi GM, Caporossi A. Corneal invasion of ocular surface squamous neoplasia after clear corneal phacoemulsification: in vivo confocal microscopy analysis. J Cataract Refract Surg. 2008;34:1038-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 48. | Xu Y, Zhou Z, Xu Y, Wang M, Liu F, Qu H, Hong J. The clinical value of in vivo confocal microscopy for diagnosis of ocular surface squamous neoplasia. Eye (Lond). 2012;26:781-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 49. | Parrozzani R, Lazzarini D, Dario A, Midena E. In vivo confocal microscopy of ocular surface squamous neoplasia. Eye (Lond). 2011;25:455-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 50. | Alomar TS, Nubile M, Lowe J, Dua HS. Corneal intraepithelial neoplasia: in vivo confocal microscopic study with histopathologic correlation. Am J Ophthalmol. 2011;151:238-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 51. | Nguena MB, van den Tweel JG, Makupa W, Hu VH, Weiss HA, Gichuhi S, Burton MJ. Diagnosing ocular surface squamous neoplasia in East Africa: case-control study of clinical and in vivo confocal microscopy assessment. Ophthalmology. 2014;121:484-491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 52. | Nanji AA, Sayyad FE, Galor A, Dubovy S, Karp CL. High-Resolution Optical Coherence Tomography as an Adjunctive Tool in the Diagnosis of Corneal and Conjunctival Pathology. Ocul Surf. 2015;13:226-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (3)] |
| 53. | Atallah M, Joag M, Galor A, Amescua G, Nanji A, Wang J, Perez VL, Dubovy S, Karp CL. Role of high resolution optical coherence tomography in diagnosing ocular surface squamous neoplasia with coexisting ocular surface diseases. Ocul Surf. 2017;15:688-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 54. | Thomas BJ, Galor A, Nanji AA, El Sayyad F, Wang J, Dubovy SR, Joag MG, Karp CL. Ultra high-resolution anterior segment optical coherence tomography in the diagnosis and management of ocular surface squamous neoplasia. Ocul Surf. 2014;12:46-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 121] [Article Influence: 9.3] [Reference Citation Analysis (1)] |
| 55. | Liu Z, Karp CL, Galor A, Al Bayyat GJ, Jiang H, Wang J. Role of optical coherence tomography angiography in the characterization of vascular network patterns of ocular surface squamous neoplasia. Ocul Surf. 2020;18:926-935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 56. | Theotoka D, Liu Z, Wall S, Galor A, Al Bayyat GJ, Feuer W, Jianhua W, Karp CL. Optical coherence tomography angiography in the evaluation of vascular patterns of ocular surface squamous neoplasia during topical medical treatment. Ocul Surf. 2022;25:8-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 57. | Dandala PP, Malladi P, Kavitha. Ocular Surface Squamous Neoplasia (OSSN): A Retrospective Study. J Clin Diagn Res. 2015;9:NC10-NC13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 58. | Barbhuiya RK, Ahmad N, Akram W. Application of Convolutional Neural Networks in Cancer Diagnosis. In: Raza K, editor. Computational Intelligence in Oncology: Applications in Diagnosis, Prognosis and Therapeutics of Cancers. Singapore: Springer, 2022: 95-109. |
| 59. | Ali AR, Li J, Kanwal S, Yang G, Hussain A, Jane O'Shea S. A Novel Fuzzy Multilayer Perceptron (F-MLP) for the Detection of Irregularity in Skin Lesion Border Using Dermoscopic Images. Front Med (Lausanne). 2020;7:297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 60. | Babenko B, Mitani A, Traynis I, Kitade N, Singh P, Maa AY, Cuadros J, Corrado GS, Peng L, Webster DR, Varadarajan A, Hammel N, Liu Y. Detection of signs of disease in external photographs of the eyes via deep learning. Nat Biomed Eng. 2022;6:1370-1383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 61. | Le D, Alam M, Yao CK, Lim JI, Hsieh YT, Chan RVP, Toslak D, Yao X. Transfer Learning for Automated OCTA Detection of Diabetic Retinopathy. Transl Vis Sci Technol. 2020;9:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 62. | Yeoh CHY, Lee JJR, Lim BXH, Sundar G, Mehta JS, Chan ASY, Lim DKA, Watson SL, Honavar SG, Manotosh R, Lim CHL. The Management of Ocular Surface Squamous Neoplasia (OSSN). Int J Mol Sci. 2022;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 63. | Sen S, Sharma A, Panda A. Immunohistochemical localization of human papilloma virus in conjunctival neoplasias: a retrospective study. Indian J Ophthalmol. 2007;55:361-363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 64. | de Koning MN, Waddell K, Magyezi J, Purdie K, Proby C, Harwood C, Lucas S, Downing R, Quint WG, Newton R. Genital and cutaneous human papillomavirus (HPV) types in relation to conjunctival squamous cell neoplasia: a case-control study in Uganda. Infect Agent Cancer. 2008;3:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 65. | Mahale A, Alkatan H, Alwadani S, Othman M, Suarez MJ, Price A, Al-Hussain H, Jastaneiah S, Yu W, Maktabi A, Deepak EP, Eberhart CG, Asnaghi L. Altered gene expression in conjunctival squamous cell carcinoma. Mod Pathol. 2016;29:452-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 66. | Palamar M, Kaya E, Egrilmez S, Akalin T, Yagci A. Amniotic membrane transplantation in surgical management of ocular surface squamous neoplasias: long-term results. Eye (Lond). 2014;28:1131-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 67. | Kieval JZ, Karp CL, Abou Shousha M, Galor A, Hoffman RA, Dubovy SR, Wang J. Ultra-high resolution optical coherence tomography for differentiation of ocular surface squamous neoplasia and pterygia. Ophthalmology. 2012;119:481-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 109] [Article Influence: 7.8] [Reference Citation Analysis (2)] |
| 68. | Abt NB, Zhao J, Huang Y, Eghrari AO. Prognostic factors and survival for malignant conjunctival melanoma and squamous cell carcinoma over four decades. Am J Otolaryngol. 2019;40:577-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 69. | Haleem A, Javaid M, Singh RP, Suman R, Rab S. Blockchain technology applications in healthcare: An overview. Int J Intell Netw. 2021;2:130-139. [DOI] [Full Text] |
| 70. | Kiire CA, Stewart RMK, Srinivasan S, Heimann H, Kaye SB, Dhillon B. A prospective study of the incidence, associations and outcomes of ocular surface squamous neoplasia in the United Kingdom. Eye (Lond). 2019;33:283-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 71. | Mirzayev I, Gündüz AK, Özalp Ateş FS, Özcan G, Işık MU. Factors affecting recurrence after surgical treatment in cases with ocular surface squamous neoplasia. Int J Ophthalmol. 2019;12:1426-1431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 72. | Singh S, Mohamed A, Kaliki S. Ocular surface squamous neoplasia: analysis based on the 8th American Joint Committee on Cancer classification. Int Ophthalmol. 2019;39:1283-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 73. | Spitzer MS, Batumba NH, Chirambo T, Bartz-Schmidt KU, Kayange P, Kalua K, Szurman P. Ocular surface squamous neoplasia as the first apparent manifestation of HIV infection in Malawi. Clin Exp Ophthalmol. 2008;36:422-425. [PubMed] |
| 74. | Responsible AI Toolkit. TensorFlow. Available from: https://www.tensorflow.org/responsible_ai. |
| 75. | Quickstart: Train an ML model with PyTorch | AI Platform Training. Google Cloud. Available from: https://cloud.google.com/ai-platform/training/docs/train-ml-model-pytorch. |
| 76. | Keras: Deep Learning for humans. Available from: https://keras.io/. |
| 77. | Gichuhi S, Sagoo MS, Weiss HA, Burton MJ. Epidemiology of ocular surface squamous neoplasia in Africa. Trop Med Int Health. 2013;18:1424-1443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 152] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 78. | Napora C, Cohen EJ, Genvert GI, Presson AC, Arentsen JJ, Eagle RC, Laibson PR. Factors associated with conjunctival intraepithelial neoplasia: a case control study. Ophthalmic Surg. 1990;21:27-30. [PubMed] |
| 79. | McClellan AJ, McClellan AL, Pezon CF, Karp CL, Feuer W, Galor A. Epidemiology of Ocular Surface Squamous Neoplasia in a Veterans Affairs Population. Cornea. 2013;32:1354-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 80. | McDonnell JM, McDonnell PJ, Sun YY. Human papillomavirus DNA in tissues and ocular surface swabs of patients with conjunctival epithelial neoplasia. Invest Ophthalmol Vis Sci. 1992;33:184-189. [PubMed] |
| 81. | Ateenyi-Agaba C, Franceschi S, Wabwire-Mangen F, Arslan A, Othieno E, Binta-Kahwa J, van Doorn LJ, Kleter B, Quint W, Weiderpass E. Human papillomavirus infection and squamous cell carcinoma of the conjunctiva. Br J Cancer. 2010;102:262-267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 82. | Scott IU, Karp CL, Nuovo GJ. Human papillomavirus 16 and 18 expression in conjunctival intraepithelial neoplasia. Ophthalmology. 2002;109:542-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 112] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 83. | Di Girolamo N. Association of human papilloma virus with pterygia and ocular-surface squamous neoplasia. Eye (Lond). 2012;26:202-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 84. | Alkatan HM, Alshomar KM, Helmi HA, Alhothali WM, Alshalan AM. Conjunctival Lesions: A 5-Year Basic Demographic Data and Clinicopathological Review in a Tertiary Eye Care Hospital. J Epidemiol Glob Health. 2022;12:25-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 85. | Asadi-Amoli F, Ghanadan A. Survey of 274 patients with conjunctival neoplastic lesions in Farabi Eye Hospital, Tehran 2006-2012. J Curr Ophthalmol. 2015;27:37-40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 86. | Kao AA, Galor A, Karp CL, Abdelaziz A, Feuer WJ, Dubovy SR. Clinicopathologic correlation of ocular surface squamous neoplasms at Bascom Palmer Eye Institute: 2001 to 2010. Ophthalmology. 2012;119:1773-1776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (3)] |
| 87. | Oellers P, Karp CL, Sheth A, Kao AA, Abdelaziz A, Matthews JL, Dubovy SR, Galor A. Prevalence, treatment, and outcomes of coexistent ocular surface squamous neoplasia and pterygium. Ophthalmology. 2013;120:445-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (2)] |
| 88. | Barros JD, Lowen MS, Moraes-Filho MN, Martins MC. Use of impression cytology for the detection of unsuspected ocular surface squamous neoplasia cells in pterygia. Arq Bras Oftalmol. 2014;77:305-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/