Published online Dec 20, 2023. doi: 10.5662/wjm.v13.i5.484
Peer-review started: July 18, 2023
First decision: September 13, 2023
Revised: September 25, 2023
Accepted: October 16, 2023
Article in press: October 16, 2023
Published online: December 20, 2023
Processing time: 155 Days and 7.1 Hours
Efficient extraction of nucleic acids and proteins (ENAP) from cells is a prerequisite for precise annotation of gene function, and has become laboratory routine for revealing the mysteries of life. However, cell samples are often from different culture dishes, resulting in inevitable experimental errors and sometimes poor repeatability.
To explore a method to improve the efficiency of ENAP, minimizing errors in ENAP processes, enhancing the reliability and repeatability of subsequent experimental results.
A protocol for the sequential isolation of RNA, DNA, and proteins from the same cultured HepG2 cells using RNAzol reagent is presented here. The first step involves culturing HepG2 cells to the exponential phase, followed by the sequential isolation of RNA, DNA, and proteins from the same cultured cells in the second step. The yield of nucleic acids and proteins is detected in the third step, and their purity and integrity are verified in the last step.
The procedure takes as few as 3-4 d from the start to quality verification and is highly efficient. In contrast to the existing kits and reagents, which are primarily based on independent isolation, this RNAzol reagent-based method is characterized by the sequential isolation of RNA, DNA, and proteins from the same cells, and therefore saves time, and has low cost and high efficiency.
The RNA, DNA, and proteins isolated using this method can be used for reverse transcription-polymerase chain reaction, polymerase chain reaction, and western blotting, respectively.
Core Tip: Sequential extraction of nucleic acids and proteins from cultured cells of the same group. Life is a way of material (mainly protein and nucleic acid) movement, and health lies in movement. Cell is the most fundamental structural and functional unit of life. Therefore, the effective isolation of nucleic acids and proteins from cells is the foundation and prerequisite for revealing the mysteries of life. However, during laboratory routine for isolation of nucleic acids and proteins, cell samples are often from different culture dishes, usually leading to inevitable experimental errors and sometimes poor repeatability. The present research tries to explore the possibility to simultaneously isolate nucleic acids and proteins from the same sample, while reducing experimental errors and ensuring consistency during experimentation. The present study established a selective protocol for sequential isolation of RNA, DNA and proteins from the same cells with the characteristics of easy operation, rapid extraction and high efficiency. RNAzol reagent was used for the sequential isolation of RNA, DNA, and proteins from the same cultured HepG2 cells, resulting in a novel protocol containing four steps. A protocol for sequential isolation of RNA, DNA and proteins was established and the procedure takes as few as 3-4 d from the start to quality verification and is highly efficient. The quality of RNA, DNA and proteins isolated through sequential isolation protocol can be used for reverse transcription - polymerase chain reaction (PCR), PCR and western blot, respectively. The present procedure is not only easy, rapid and high efficient, but also economical and practical, especially for researchers in developing and underdeveloped countries.
- Citation: Cui YY. Sequential extraction of RNA, DNA and protein from cultured cells of the same group. World J Methodol 2023; 13(5): 484-491
- URL: https://www.wjgnet.com/2222-0682/full/v13/i5/484.htm
- DOI: https://dx.doi.org/10.5662/wjm.v13.i5.484
The essence of life relies on molecular kinetics, which primarily involves the interactions of nucleic acids (DNA and RNA) and proteins. Therefore, the extraction of nucleic acids and proteins is crucial for deciphering the secrets of life. It is becoming increasingly important for researchers in the post-genomic era to efficiently isolate nucleic acids and proteins of high purity and integrity from cultured cells. Various commercial kits are available for the isolation of nucleic acids and proteins, using various methods[1]. Recently, methods for RNA[2], DNA[3] and protein[4,5] micro-isolation have been updated, respectively, and simultaneous isolation of RNA and DNA or protein has also been reported[6,7], but there has been no reported method for simultaneous isolation of RNA, DNA, and protein from cultured cells of the same group. Methods for the independent extraction of nucleic acids and proteins from different groups of cultured cells comprise several steps, are expensive, and time-consuming; these factors affect the productivity, purity, and integrity of the isolated samples. Most importantly, it is very difficult to ensure that the number of cells, cell growth status, and metabolic status of the cultured cells are consistent across the different groups, which inevitably increases the chances of experimental errors between earlier and later experiments. Although the complete elimination of errors is not achievable during experimentation, it is essential to minimize errors as far as possible. The quality of the RNA and proteins isolated from different groups of cultured cells could be a key factor responsible for the inconsistencies in gene expression data obtained by reverse transcription (RT)-polymerase chain reaction (PCR) and western blotting that are often observed. By referring to related literature[8-10] and repeated experimentation, the present study established a relatively rapid procedure for the sequential extraction of RNA, DNA, and proteins from the same group of cultured cells. The method described herein is not only easy and inexpensive, but also has high reproducibility, comparability, and credibility, and ensures consistency during experimentation.
The following reagents were used in this study for the successive extraction of nucleic acids and proteins: RPMI 1640 (HyClone), penicillin-streptomycin (10000 units/mL penicillin and 10000 μg/mL streptomycin; Invitrogen), HEPES (Sigma, United States), fetal bovine serum (FBS; Sijiqing, China), RNAzol (Cohen-Bio Corp., Beijing, China, lot no. NA6111), sodium dodecyl sulfate (SDS; Amresco), agarose (Fluka, Spain), diethyl pyrocarbonate (DEPC; Sigma), acrylamide (BBI, Canada), bis-acrylamide (BBI), tetramethylethylenediamine (TEMED; Sigma), ammonium persulfate (Sigma,), guanidine hydrochloride (BBI, Canada), Tris (Sigma), glycine (Sigma), ethidium bromide (E.B; Sigma), MOPS (Serva, Sino-American Biotechnology Co.), PRO-STAINTM protein marker II (SBS Genetech Co., Beijing, China), horseradish peroxidase-anti-glyceraldehyde-3-phosphate dehydrogenase (HRP-anti-GAPDH; Kangchen Bio-tech, Shanghai, China), and sediment type mono-ingredient TMB substrate solution (PA108-01, Tiangen Biotech Co., Beijing, China).
The following primers were used in this study: c-Myc: Forward: 5′-AGCAAACCTCCTCACAGC-3′, and reverse: 3′-GATGCCTTGAGAACACGC-5′ (GenBank accession number: NM_002467).
The following equipment was used for the sequential extraction of nucleic acids and proteins: CO2 water jacketed incubator (Thermo Forma MODEL 3111, series II, HEPA FILTER, Forma Scientific Inc.), inversion microscope (XDS-1B, COIC), an agarose gel electrophoresis apparatus (DYY-III-4, LiuYi Corp. Beijing), SDS-polyacrylamide gel electrophoresis (PAGE) apparatus (POWER-PAC200/300, Bio-Rad), a MultiMage™ Light Cabinet (Alpha Innotech Corporation), and a ultraviolet spectrometer (Ultrospec®2100 Pro, Amersham Pharmacia Biotech).
Duration: 1-3 d, depending on the cell line used.
The cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 10 mmol/L HEPES at pH 7.4, and maintained at 37 °C in a humidified incubator with 5% CO2. When the cells reached a confluence of 75%, they were detached with 0.25% trypsin and subsequently seeded into 24-well microtiter plates at a density of 104 cells /mL with 2 mL complete culture media /well, and cultured in a 5% CO2 incubator for 24, 48, 72, and 96 h. The cells were collected from three wells at each time point and counted with a hemocytometer. The mean number of cells calculated from the three wells represented the у-axis value, and the time point was represented on the x-axis of a cell growth curve, which was analyzed to identify the exponential growth phase of the cells.
Duration: Approximately 1 h.
Homogenization: The cells were directly lysed in the exponential phase of growth in a culture dish by adding 1 mL RNAzol reagent to a dish of diameter 3.5 cm, and the cell lysate was pipetted several times for homogenization.
The homogenized samples were incubated for 5 min at room temperature (RT, 25 °C) to allow the complete dissociation of nucleoprotein complexes.
The samples were centrifuged at 11000 × g for 10 min at 4 °C, following which the clear supernatant solution was transferred to a fresh 1.5 mL Eppendorf tube.
A 0.2 mL aliquot of chloroform was added to the Eppendorf tube and the sample tubes were securely capped. The tubes were vigorously shaken by hand for 15 s and incubated at RT for 3 min.
The samples were centrifuged at 11000 × g for 15 min at 4 °C. Following centrifugation, the mixture separated into a lower, phenol-chloroform phase, an interphase, and an upper colorless aqueous phase. The RNA remained exclusively in the aqueous phase, the DNA remained in the interphase, and the proteins were retained in the lower organic phase. The volume of the aqueous phase was approximately 60% of the volume of RNAzol reagent used for homogenization. The aqueous, interphase, and organic phases were collected in fresh 1.5 mL Eppendorf tubes, and the tubes containing the interphase and organic phase were stored at 4 °C for the isolation of DNA and proteins.
A 0.5 mL aliquot of isopropyl alcohol was added to the tube containing the aqueous phase, mixed evenly, and allowed to stand at room temperature for 10 min.
The mixture was centrifuged at 11000 × g for 10 min at 4 °C and the supernatant was discarded. The RNA precipitate, often invisible before centrifugation, formed a gel-like pellet on the sides and bottom of the tube.
At least 1 mL of 75% ethanol was added for washing the RNA pellet, once.
The sample was mixed by vortexing and subsequently centrifuged at a speed < 7500 × g for 5 min at 4 °C. The supernatant obtained after centrifugation was discarded.
The RNA pellet was air-dried for 5-10 min. The RNA was dissolved in an appropriate volume of RNase-free water by pipetting the solution a few times and incubating for 10 min at 55-60 °C. A 2 μL aliquot of the RNA solution was diluted 200-fold for detection of absorbance (A) at 260 nm, and the remnant was stored at - 80 °C until further use.
Duration: Approximately 1 h.
Any remaining aqueous phase was removed from the interphase layer and discarded, and 0.3 mL of 100% ethanol was added per milliliter of RNAzol reagent used for the initial homogenization of the interphase and phenol phases. The samples were mixed by inversion and allowed to stand at room temperature for 3 min.
The DNA was sedimented by centrifugation at a speed less than 2000 × g for 5 min at 4 °C. The phenol-ethanol supernatant was then transferred to a fresh tube and stored at 4 °C for protein isolation.
A 0.3 mL aliquot of 0.1 M sodium citrate-10% ethanol solution was added to wash the DNA pellet, at least twice. During each wash, the DNA pellet was kept in the washing solution for 30 min at room temperature (with periodic mixing) and centrifuged at 2000 × g for 5 min at 4 °C.
The DNA pellet was suspended in 75% ethanol (1.5-2 mL of 75% ethanol per mL of RNAzol reagent), stored for 20 min at room temperature (with periodic mixing), and centrifuged at 2000 × g for 5 min at 4 °C.
The DNA was air-dried for 5-15 min in an open tube. The DNA isolated from 106 cells was dissolved by adding 20-60 μL of 8 mmol/L NaOH, such that the concentration of the isolated DNA reached 0.2-0.3 μg/μL.
Duration: Approximately 1 h.
For protein isolation, 1.5 mL of isopropanol was added to the previously obtained phenol-ethanol supernatant (approximate volume: 0.8-1 mL of RNAzol reagent), and allowed to stand at room temperature for 10 min.
The protein precipitate was sedimented by centrifugation at 12000 × g for 10 min at 4 °C, and the supernatant was discarded.
A 2 mL solution of 0.3 M guanidine hydrochloride in 95% ethanol was added to wash the protein pellet, thrice. During each wash cycle, the protein pellet was kept in the wash solution for 20 min at room temperature (15-30 °C) and centrifuged at 7500 × g for 5 min at 4 °C. After the final wash, the protein pellet was vortexed in 2 mL ethanol, stored in ethanol for 20 min at room temperature, and finally centrifuged at 7500 × g for 5 min at 4 °C.
The protein pellet was air-dried for 5-10 min and dissolved in 50 μL of 1% SDS by pipetting. Notably, the complete dissolution of the protein pellet might require incubating the sample at 50 °C. Any insoluble material was sedimented by centrifugation at 10000 × g for 10 min at 4 °C, and the supernatant was transferred to a fresh tube. A 1 μL aliquot of the sample was subsequently used for detecting the concentration and purity of the isolated proteins, and 10 μL of the sample was used for western blotting. The remainder was stored at −20 °C for future use.
The expected yields of RNA, DNA, and proteins from 1 × 106 cultured HepG2 cells are 5-10 μg, 4-7 μg, and 10-12 μg, respectively. Nucleic acids and proteins have an A280/A260 ≥ 1.8 when diluted with Tris-EDTA buffer (TE; 10 mmol/L Tris, 1 mmol/L EDTA, pH 8.0) and 0.1% SDS, respectively.
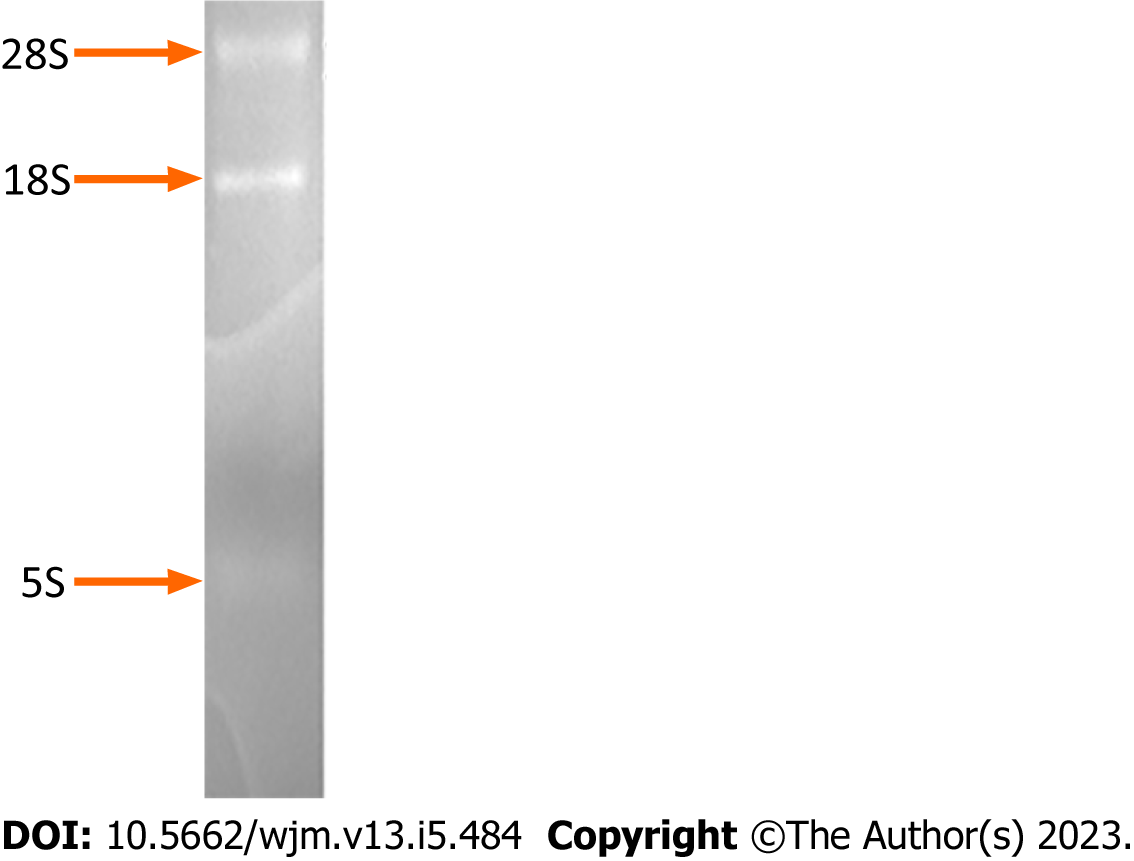
Electrophoresis of the isolated RNA on a formaldehyde-denaturing agarose gel and subsequent staining with ethidium bromide revealed discrete bands of high molecular weight, corresponding to RNA molecules of size 7-15 kb (mRNAs and hnRNAs), two predominant ribosomal RNA bands of size ~5 kb and ~2 kb (28S and 18S rRNAs, respectively), and a low molecular weight RNA molecule of 0.1-0.3 kb (tRNA, 5S) (Figure 1).
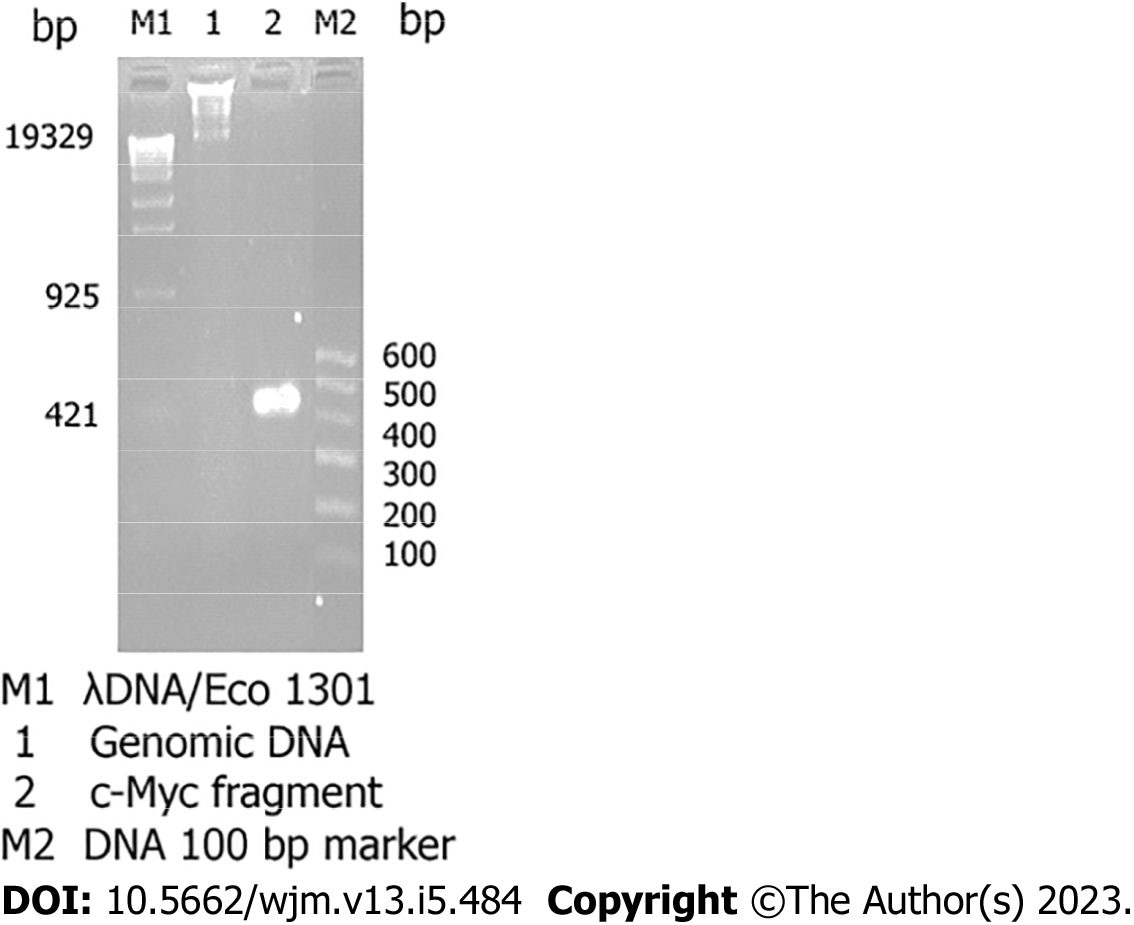
The total RNA obtained using this method is free from contamination with protein and DNA and can be used for molecular cloning (RT-PCR) (Figure 2).
The DNA isolated using this method can be used for the detection of integrated foreign genetic materials by using PCR and restriction endonucleases.
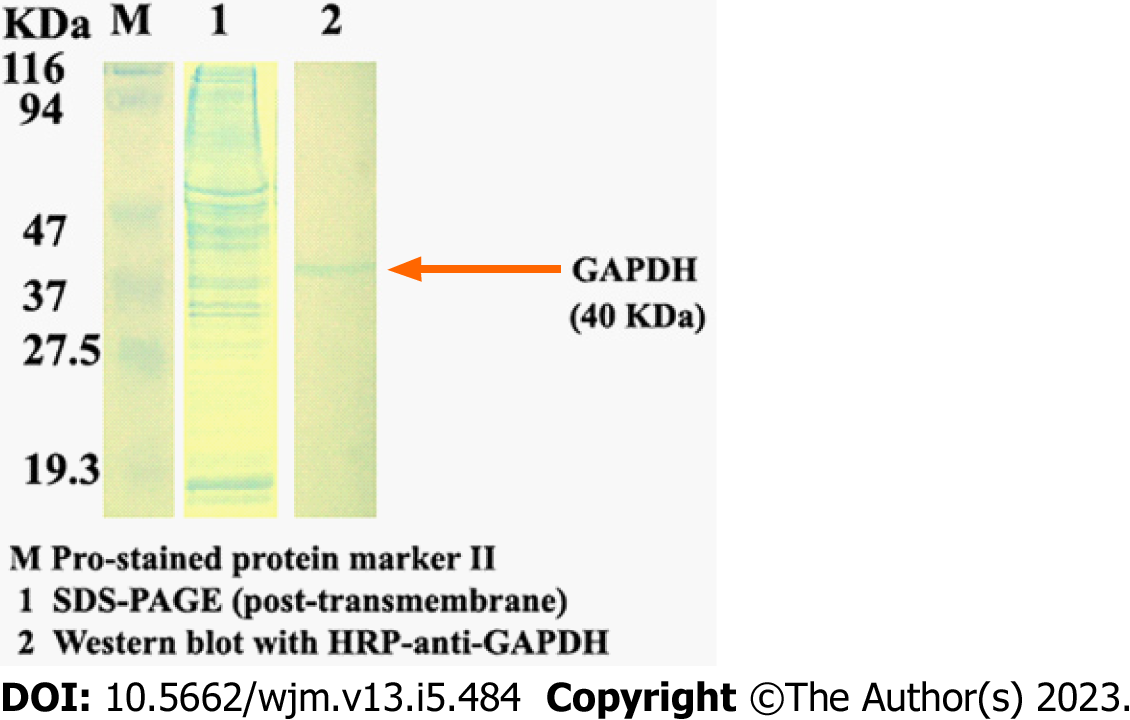
The resulting protein isolated by this method can be analyzed for the presence of specific proteins by western blotting (Figure 3).
The cells should be lysed directly in a culture dish with appropriate RNAzol reagent only when they are in the exponential phase of growth, i.e. only cells in the exponential phase of growth should be used for isolation.
Disposable gloves should be worn always, and sterile, disposable plasticware and automatic pipettes should be reserved for RNA isolation. The glassware and plasticware should be kept RNase-free during RNA isolation to protect the RNA from contamination and degradation by RNases.
The isolated RNA and DNA samples should not be dried by centrifugation under vacuum.
Understanding the functions of genes in the postgenomic era is crucial for deciphering cellular mechanisms. Therefore, the development of effective methods of cell culture and novel techniques for the isolation of RNA, DNA, and proteins from cultured cells, especially the microextraction of nucleic acids and proteins from < 106 cultured cells, have become increasingly important. The protocol described herein offers such a method for isolating nucleic acids and proteins with relative rapidity and efficiency. The results demonstrate that the procedure allows the sequential isolation of RNA, DNA, and protein with high purity and integrity. The advantage of this method is that it allows the almost synchronous isolation of nucleic acids and proteins from the same cultured cells, which not only saves time, money, manpower, and material resources, but also preserves the identity of the isolated materials and enhances the reproducibility and reliability of the experimental results.
RNAzol is a ready-to-use reagent that is used for the isolation of total RNA from cells. The reagent consists of a monophasic solution of phenol and guanidine isothiocyanate, and the RNAzol-based technique of RNA isolation is superior to the single-step RNA isolation method developed by Chomczynski and Sacchi[8,9]. RNAzol maintains the integrity of the RNA during cell lysis and dissolution of cellular components. The addition of chloroform followed by centrifugation separates the solution into an aqueous phase and an organic phase, and the RNA remains exclusively in the aqueous phase. The RNA is recovered by precipitating the aqueous phase with isopropyl alcohol. The DNA in the interphase layer can then be isolated by precipitation with ethanol, and the proteins can be isolated from the organic phase by an additional precipitation step with isopropyl alcohol[4,10].
In the classical isolation methods, RNA, DNA, and proteins are extracted independently, which requires the preparation of a triplet group of cells as well as several reagents that need to be added separately during isolation. These protocols involve numerous steps and are time-consuming. In these conditions, RNA can easily be contaminated and degraded by extraneous RNases. Therefore, the operational difficulty of the classical isolation methods is higher and the chances of successful isolation are decreased. Additionally, the various commercially available kits for the extraction of nucleic acids are expensive. RNAzol reagent facilitates the sequential isolation of RNA, DNA, and proteins from the same cultured cells, and preserves the identity of the isolated materials. The method developed herein has several advantages, including good comparability and reproducibility, a simple protocol, short duration of experimentation, improved work efficiency, reduced chance of RNA degradation, and it does not require the use of proteinase inhibitors for isolating single proteins. The quality of the isolated RNA, DNA, and proteins was validated by formaldehyde-denaturing agarose gel electrophoresis, RT-PCR, SDS-PAGE, and western blotting. And the findings reveal that the isolated nucleic acids and proteins can be used in molecular biology studies. Of note, this protocol is not suitable for lifelong cells without proliferative ability, e.g. neural cells and myocardial cells, or cells with relatively weak proliferative ability, e.g. stem cells with relatively small numbers.
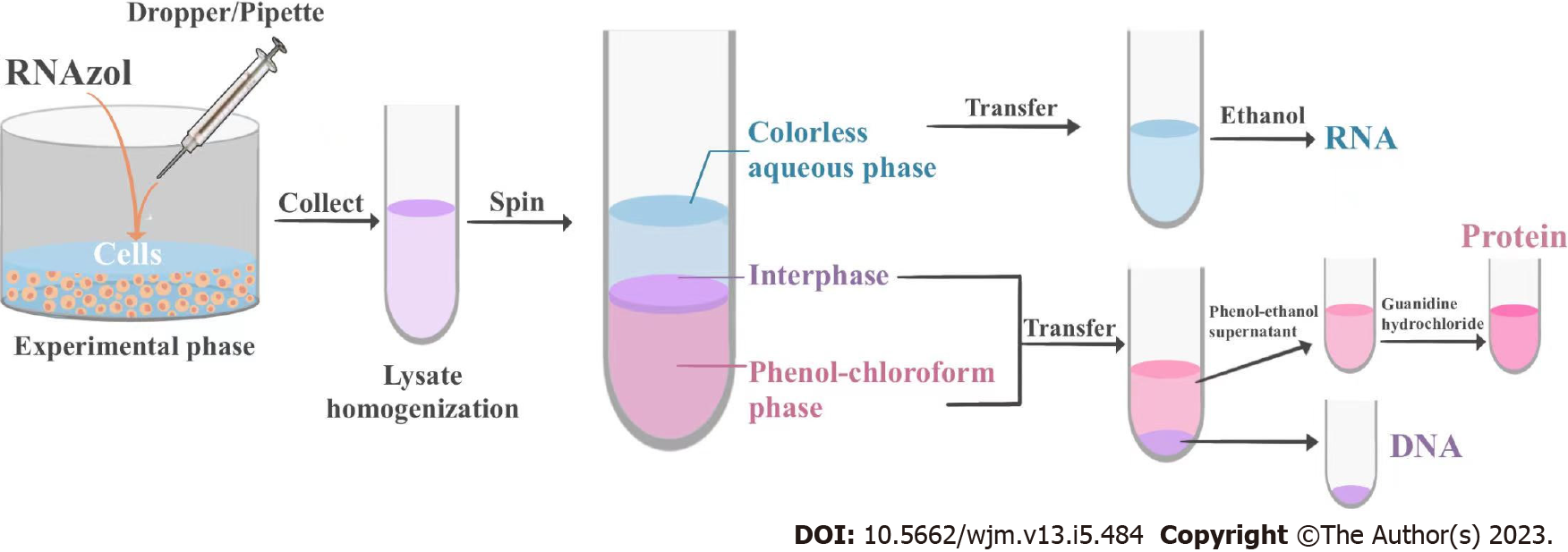
Together, the present study describes a novel protocol for the sequential micro-extraction of RNA, DNA, and proteins from the same cells (Figure 4). The procedure has easy operation, allows rapid isolation, has high efficiency, and is economical and practical, especially for researchers in developing countries.
Life is a way of material (mainly protein and nucleic acid) movement and health lies in movement. Cell is the most fundamental structural and functional unit of life, therefore, the effective isolation of nucleic acids and proteins from cells is the foundation and prerequisite for revealing the mysteries of life. However, during laboratory routine for isolation of nucleic acids and proteins, cell samples are often from different culture dishes, usually leading to inevitable experimental errors and sometimes poor repeatability.
The present research tries to explore the possibility to simultaneously isolate nucleic acids and proteins from the same sample, while reducing experimental errors and ensuring consistency during experimentation.
The present study established a selective protocol for sequential isolation of RNA, DNA and proteins from the same cells with the characteristics of easy operation, rapid extraction and high efficiency.
RNAzol reagent was used for the sequential isolation of RNA, DNA, and proteins from the same cultured HepG2 cells, resulting in a novel protocol containing four steps.
A protocol for sequential isolation of RNA, DNA and proteins was established and the procedure takes as few as 3-4 d from the start to quality verification and is highly efficient.
The quality of RNA, DNA and proteins isolated through sequential isolation protocol can be used for reverse transcription (RT) - polymerase chain reaction (PCR), PCR and western blot, respectively.
The present procedure is not only easy, rapid and high efficient, but also economical and practical, especially for researchers in developing and underdeveloped countries.
I thank my laboratory mates for their sincerely help.
| 1. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 3th ed. New York: Cold Spring Harbor Laboratory Press, 2001: 131-145. |
| 2. | Alabi T, Patel SB, Bhatia S, Wolfson JA, Singh P. Isolation of DNA-free RNA from human bone marrow mononuclear cells: comparison of laboratory methods. Biotechniques. 2020;68:159-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | Tripodi P, Festa G. High-Throughput DNA Isolation in Vegetable Crops for Genomics Applications. Methods Mol Biol. 2021;2264:47-53. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 4. | Azzimato V. Kupffer Cell Protein Isolation and Detection by Western Blot. Methods Mol Biol. 2020;2164:21-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 5. | Blanco-Fernandez J, Jourdain AA. Two-Step Tag-Free Isolation of Mitochondria for Improved Protein Discovery and Quantification. J Vis Exp. 2023;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 6. | Grima N, Henden L, Watson O, Blair IP, Williams KL. Simultaneous Isolation of High-Quality RNA and DNA From Postmortem Human Central Nervous System Tissues for Omics Studies. J Neuropathol Exp Neurol. 2022;81:135-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 7. | Melnik S, Caudron-Herger M, Brant L, Carr IM, Rippe K, Cook PR, Papantonis A. Isolation of the protein and RNA content of active sites of transcription from mammalian cells. Nat Protoc. 2016;11:553-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40518] [Cited by in RCA: 39209] [Article Influence: 1005.4] [Reference Citation Analysis (0)] |
| 9. | Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc. 2006;1:581-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1288] [Cited by in RCA: 1419] [Article Influence: 71.0] [Reference Citation Analysis (0)] |
| 10. | Xiong RP, Liu P, Zhou YG, Chen XY, Wang H. Extraction of trace total RNA and protein from small quantity cultured cells. Chin J Biochem Mol Biol. 2003;19:256-260. |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medical laboratory technology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Castelucci P, Brazil; I-Shiang Tzeng, Taiwan; Sheykhhasan M, Iran; Timotius Ivan Hariyanto, Indonesia S-Editor: Li L L-Editor: A P-Editor: Zhang YL