INTRODUCTION
Precision medicine is the new frontier of healthcare and medical research, and it has been mainly implemented in oncology. Precision oncology can be defined as an approach for treatment and prevention of cancer with the individual variability in genes, environment, and behavior taken into account. Accurate risk assessment, prevention, detection, and treatment of cancer tailored to the individual are major challenges in clinical oncology. Despite various advances in basic, translational, and clinical cancer research, the incidence and mortality rates of malignant diseases have remained high[1,2]. Artificial intelligence (AI), a field of applied computer science, has shown promising potential of accelerating towards the goal of precision oncology. Application of AI in oncology involves integrative analysis of “big cancer data” such as digitized images, multi-omics, clinical datasets, and population health.
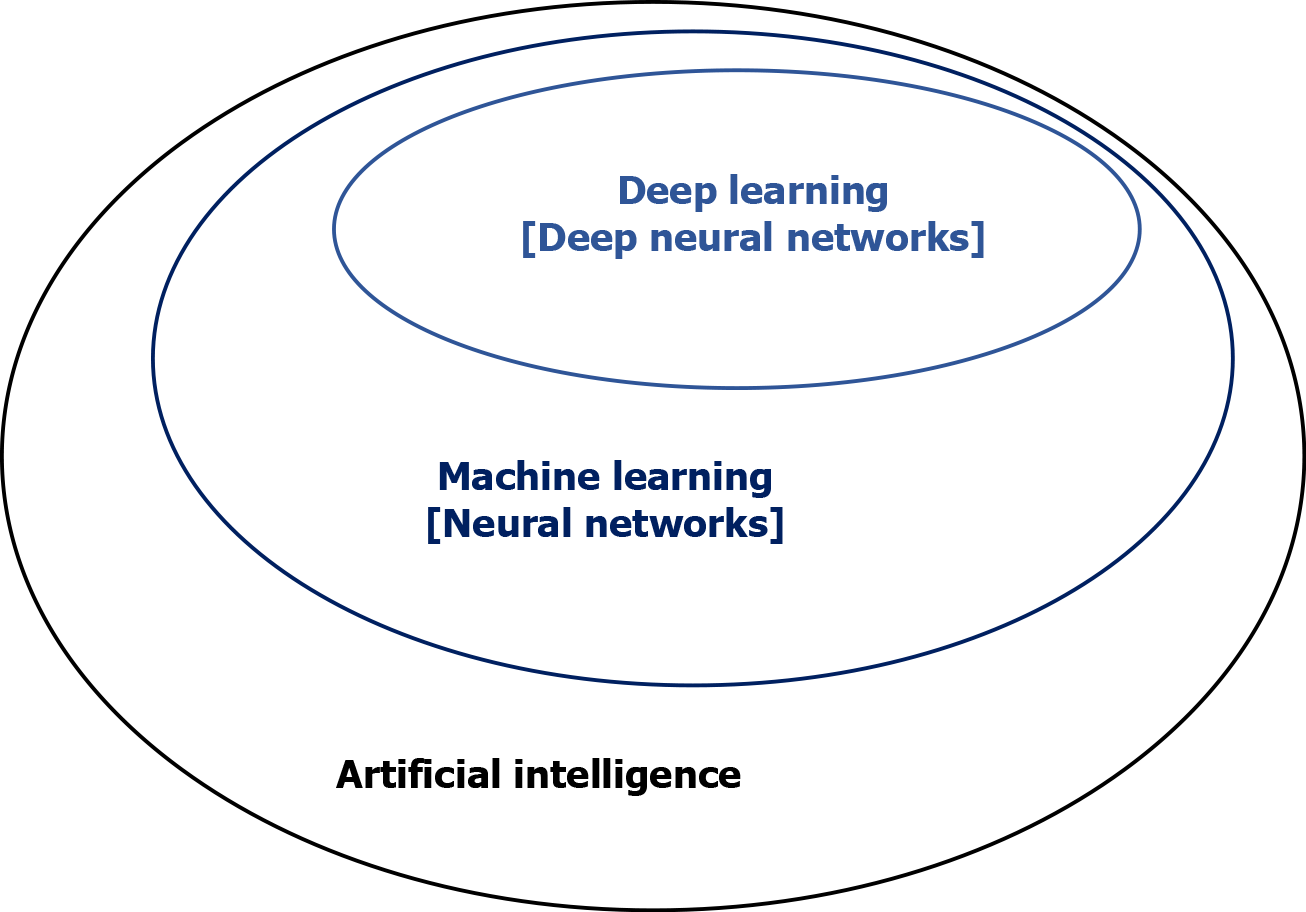
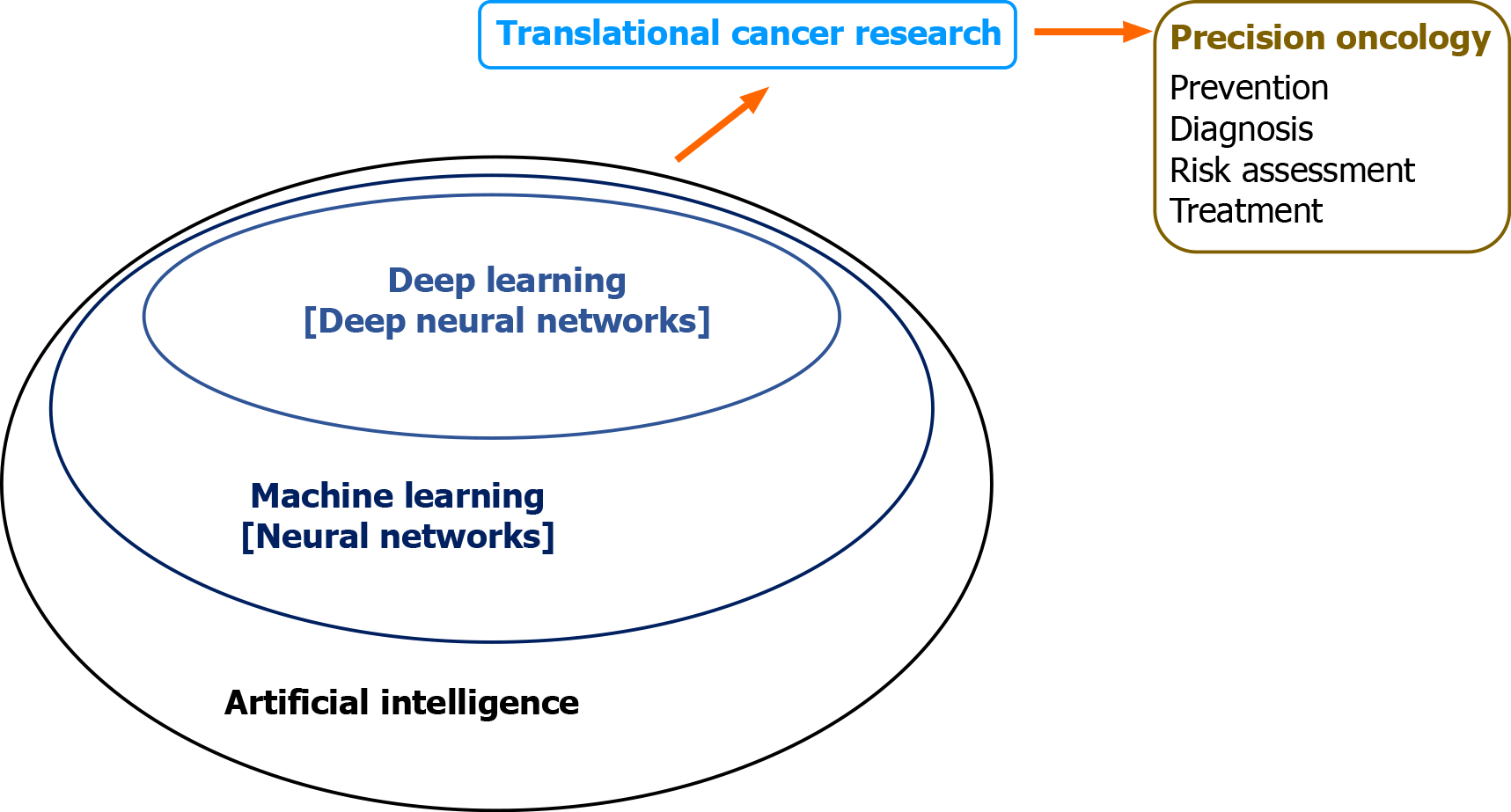
With the advent of electronic health records, bio-banking, multi-omics, and digitized radiographic and histological images, we have entered the era of big data and team science. AI has emerged as a powerful technology that will transform healthcare by multi-disciplinary research to capture and analyze large pools of data[3,4]. Application of AI in translational cancer research has shown its potential for advancing diagnosis, prognostication, and treatment[5]. This is accomplished by integration and analysis of large data sets and generating algorithms-based prediction models. Machine learning (ML) is a branch of AI that applies statistical methods to detect patterns within datasets[6]. Deep learning (DL), characterized by deep artificial neural network, is a sub-branch of ML that utilizes the capability of multi-layered networks[7] (Figures 1 and 2). Application of ML and DL approaches has been demonstrated to advance translational cancer research in various aspects. These include detection and classification of tumor subtypes, diagnosis of cancer, assessment of cancer risk, prediction of clinical outcomes, discovery of cancer biomarkers, repurposing of drugs for cancer treatment, and predicting drug response of tumors.
Figure 1 Artificial intelligence, machine learning, and deep learning.
Artificial intelligence is a field of applied computer science that mimics human cognition to complete a task. Machine learning is a branch of artificial intelligence that manually extract features from input data to create a model that categorizes the object. Neural network is a set of machine-based learning algorithms to learn labeled datasets and perform classification tasks, and it comprises an input layer, a hidden layer of interconnected nodes, and an output layer. Deep learning is a machine learning technique that uses neural network architectures, and the term “deep” refers to the number of hidden layers (more than three) in the neural network.
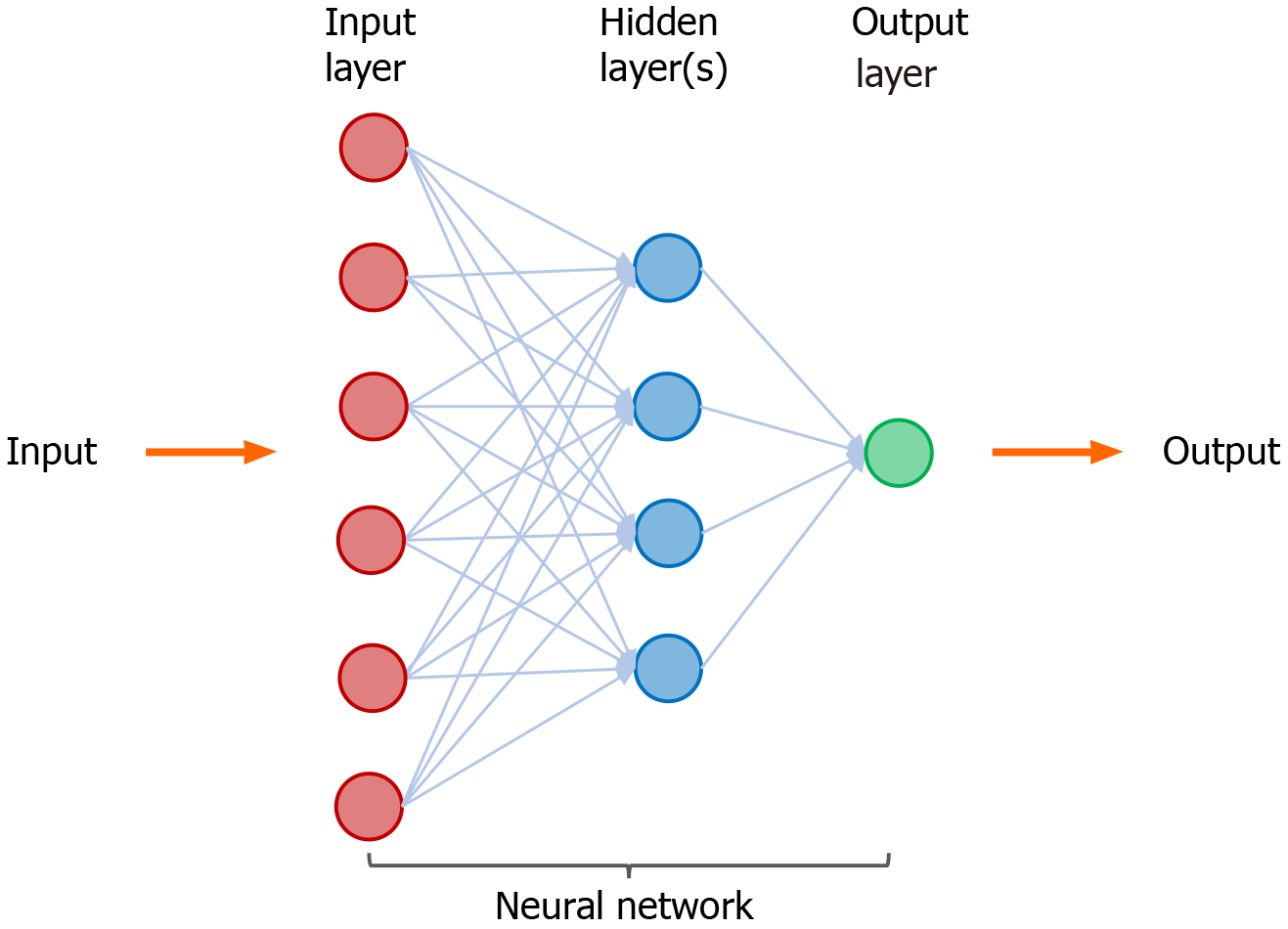
Figure 2 A neural network in machine learning.
Neural network is a set of machine-based learning algorithms, and it comprises an input layer (red circles), a hidden layer of interconnected nodes (blue circles), and output layer (green circle). The function of a neural network is to extract and process features from labeled datasets and perform classification tasks. A representative hidden layer of interconnected nodes (blue circles) is shown. A deep learning node (blue circle) is a computational unit that combines input data with weights (assigned significance) and generate an output layer. For deep learning, a neural network contains more than three hidden layers.
ML IN RADIOLOGICAL IMAGES OF CANCER
Classification and early detection of cancer are crucial for accurate diagnosis and treatment with curative intent. In “radiomics”, the data based on algorithm for extracting and analyzing features from medical images enable improvement of accuracy of cancer diagnosis, prognostication, and clinical prediction[8]. Advances have been made in research of DL with convolutional neural networks (CNN), an algorithm to process and differentiate images, in cancer imaging and help facilitate accurate classification and detection of cancer[9] (Figure 3).
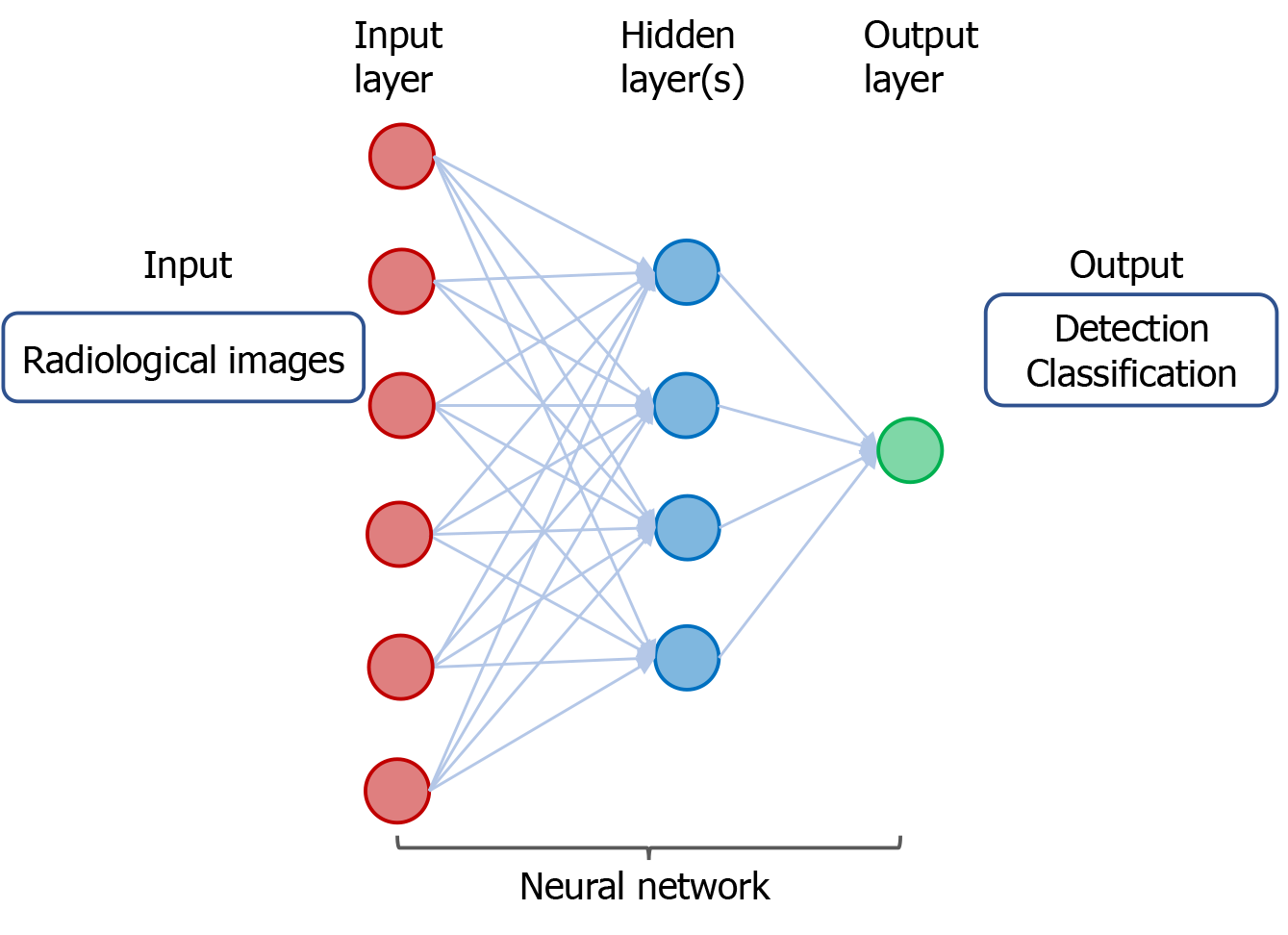
Figure 3 A neural network in deep learning of radiological images.
The input consists of data derived from radiological images from individuals with cancer or without cancer. The output includes detection and classification of tumors.
DL with CNN and its variants has been applied for classification and detection of cancer in different organs. Several studies using DL in radiological images as input data are described as follows. Using a dataset of about 130000 clinical images of skin lesions, a trained CNN is capable of a dermatologist-level classification of keratinocyte carcinoma and malignant melanoma[10]. As shown in a systematic review of eleven studies, CNN enables accurate diagnosis of hepatocellular carcinoma by recognizing specific features in computed tomographic (CT) or magnetic resonance images[11]. Based on retrospective datasets of 2652 digital mammography, 653 of which showed malignancy, the AI system using DL CNN algorithms to detect calcifications and soft tissue lesions showed an accuracy for detection of breast cancer comparable to an average breast radiologist[12]. Using two independent datasets for training and validating the AI algorithm, the accuracy of a DL-based model for screening breast cancer by mammography is superior to that of expert radiologists, with the area under the receiver operating characteristic curve (AUC-ROC) for the AI system greater than that for the average radiologist by 11.5%[13]. By combining 3-dimensional deep CNN with cloud computing for analysis of the datasets of lung nodules on chest CT imaging, lung cancer can be accurately detected with a sensitivity of 98.7% at 1.97 false positives per scan[14].
Results of these studies demonstrate the power of AI using DL CNN algorithms for accurately detecting cancer in different organs (Figure 3). Future investigation by clinical trials is indicated to improve the accuracy and efficiency of cancer detection using the AI systems. Multi-disciplinary and coordinated research efforts are necessary to determine how the DL-based models can be potentially integrated into clinical practice.
ML IN HISTOLOGICAL IMAGES OF CANCER
Microscopic analysis of tumor histopathology with immunohistochemistry has been the standard practice for diagnosis and grading of cancer. With the advent of scanning technology, digitization of whole slide images of biopsied or resected tumor specimens has enabled computer-assisted analysis to improve accuracy and efficiency of diagnosis. ML/DL for analysis of digitalized images of tumor histopathology has been demonstrated to have potential for improving diagnosis of cancer, identifying tumor and lymph node metastasis, predicting genetic mutation and clinical outcomes[15] (Figure 4).
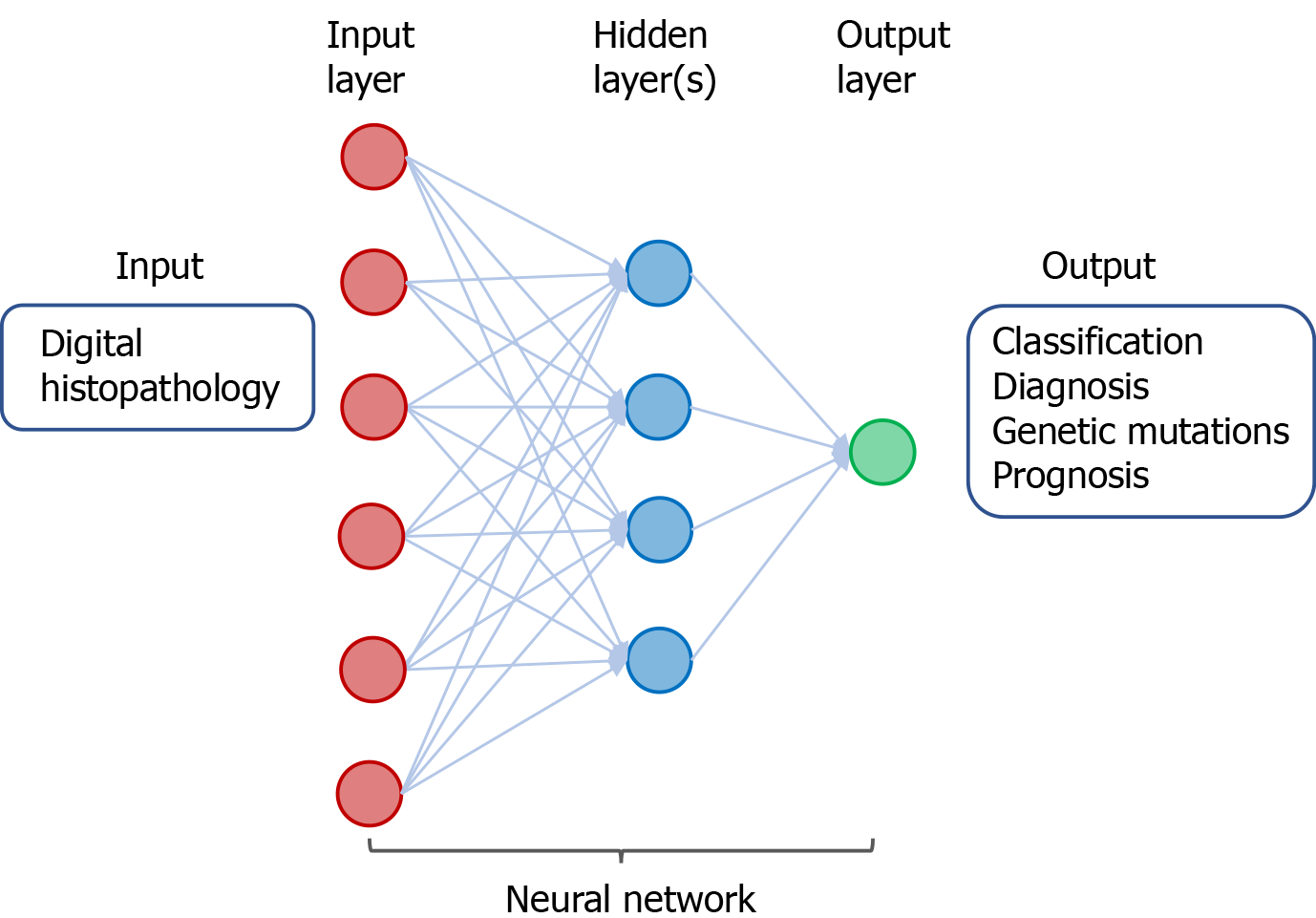
Figure 4 A neural network in deep learning of digital histopathology.
The input consists of digital histopathological data derived from individuals with or without cancer. The output includes classification and diagnosis of cancer as well as predicting genetic mutations and prognosis of patients with cancer.
Here are a few examples that illustrate the power of ML in digitized images of cancer for improving accuracy of pathological diagnosis. Using DL with CNN technique for analysis of biopsied tissue specimens, prostate cancer as well as micro- and macro-metastases of breast cancer in sentinel lymph nodes could be automatically identified without the need for immunohistochemistry[16]. Similarly, using DL-based approaches to train a CNN to discriminate tumor from normal tissue, metastatic breast cancer could be automatically detected in images of biopsied sentinel lymph nodes[17,18].
Moreover, DL-based algorithms can be trained to analyze histopathological images and predict mutation and clinical outcomes. A deep CNN was trained to analyze whole slide images obtained from The Cancer Genome Atlas (https://portal.gdc.cancer.gov). The DL method could automatically classify the tissues as lung adenocarcinoma, squamous cell carcinoma, or normal lung tissue. In addition, the trained CNN could accurately predict some of the commonly mutated genes in lung adenocarcinoma including EGFR, KRAS, TP53, FAT1, SETBP1, and STK11[19]. A novel DL-based approach to train a deep network was used to analyze digitized tissue microarray specimens of colorectal cancer from 420 patients, along with their clinicopathological features and clinical outcomes. Results of this study show that DL-based prediction of 5-year disease specific survival is superior to that by visual evaluation of histology by expert pathologists[20].
DL with CNN approaches have shown promising potential for improving digitized histopathology-based diagnostics (Figure 4). In order to apply for diagnosis and classification of cancer in clinical practice, the utility of ML/DL in analyzing digital tumor histopathology will need to be assessed and validated in prospective clinical trials involving a large number of patients. Furthermore, combination of ML in digitized histopathology with other datasets such as tumor omics can enhance the predictive capability and accuracy.
ML IN MULTI-OMICS DATASETS OF CANCER
Research on molecular characterization of tumors has generated a wealth of data on genetic and epigenetic alterations that control carcinogenesis[21]. Tissue-based omics have yielded tremendous number of clinically useful cancer biomarkers and targets. These include data derived from genomics, epigenomics, transcriptomics, proteomics, metabolomics, phenomics, and metagenomics [Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov)][22]. These omics data have been used to classify tumor types, identify and develop cancer biomarkers, and drug discovery and development. Moreover, ML/DL may help improve the efficiency and accuracy omics-based therapeutic strategies[23](Figure 5).
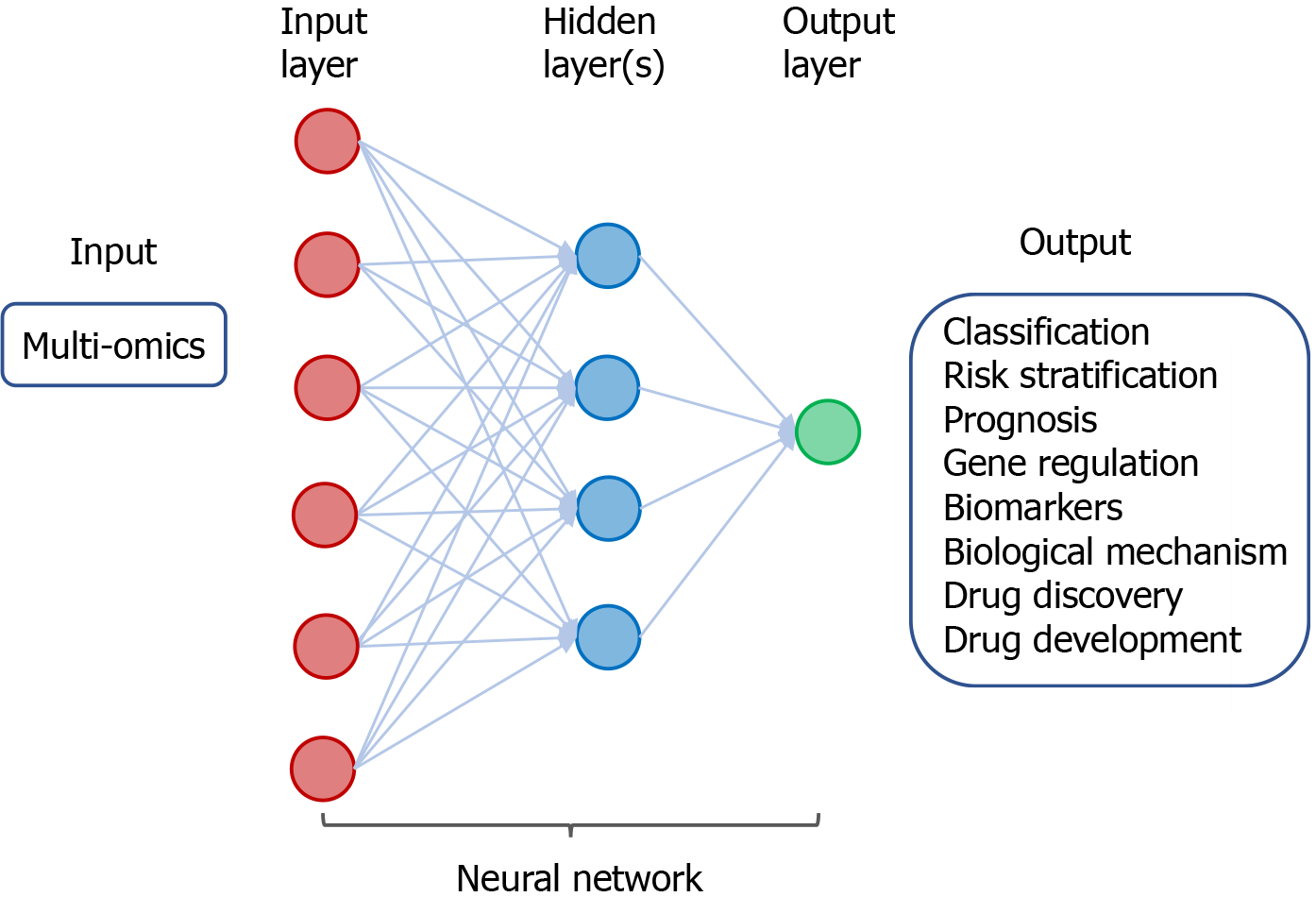
Figure 5 A neural network in deep learning of cancer-derived multi-omics.
The input consists of multi-omics data derived from various cancers. The output includes classification and risk stratification of cancer, predicting prognosis, and investigating gene regulation/biomarkers/biological mechanism, as well as drug discovery and development.
DL algorithms for analysis of omics data have been demonstrated to facilitate classification and detection of cancer as well as stratification of risk in patients with cancer. Using a DL approach, termed Stacked Denoising Autoencoder, to extract features from RNA sequencing (RNA-seq) expression in The Cancer Genome Atlas (TCGA) database, breast tissues can be classified into cancer or non-cancer and the involved genes can be identified as potential cancer biomarkers[24]. By application of CNN for analysis of RNA-seq data in Pan-Cancer Atlas, tissue samples have been classified with accuracy into 33 different types of cancer[25]. Besides gene expression data, DL analysis of epigenetics data particularly DNA methylation in the context of CpG islands has also been shown to classify cancer types. A deep neural network (DNN) was developed to extract the deep features of DNA methylation data, and this method can differentiate patients with breast cancer from healthy individuals[26]. Similarly, a CNN-based DL model can classify different types of cancer by analysis of the patterns of DNA methylation[27]. Furthermore, an advanced DNN-based model, DeepGene, was developed to analyze somatic point mutation data, and it was demonstrated to improve classification of 12 selected types of cancer[28]. Recently, the power of DL and traditional ML methods in cancer classification using TCGA datasets was compared, and results of the study indicate that the DL method, termed Multi-Layer Perceptions, outperforms the other approaches in discrimination of samples with cancer from non-cancer[29].
In addition to classification of cancer types, DL and ML have been exploited to predict patient prognosis and investigate gene regulation. By DL-based analysis of multi-omics data, including RNA-seq, microRNA sequencing (miRNA-seq), and DNA methylation data, patients with hepatocellular carcinoma can be classified into subgroups with difference in survival[30]. DL autoencoder algorithm for analysis of multi-omics datasets comprising mRNA, miRNA, and DNA methylation from TCGA can also predict the survival subtypes of patients with urinary bladder cancer[31]. Besides, ML-based integrative analysis of multi-omics data on the cloud has been demonstrated to improve the accessibility and productivity of cancer research for discovery of gene regulatory subnetwork, analysis of disease subtype, analysis of survival, prediction of clinical outcome, and visualization of multi-omics results[32].
By application of DL for integrative analysis of omics data, prediction models can be generated for discovering biomarkers and repurposing drugs[33,34]. In a proof-of-principle study using transcriptomic profiles of cancer cells treated with a variety of drugs at different concentrations, DNNs were trained to classify those drugs into therapeutic categories[35]. Using chemical and genetic information, a DL approach was shown to model and predict synergistic actions of novel combinations of anti-cancer drugs[36]. A DNN-based framework, termed PADME (Protein And Drug Molecule interaction prediction), was developed to predict drug-target interaction with input of information on compounds and protein[37].
Application of ML/DL in analyzing multi-omics has shown utility for classification of cancer and its risk stratification prognosis, demonstrated the power of investigating gene regulation, biomarkers, and biological mechanism, and created ample opportunity for drug discovery and development (Figure 5). Emerging studies have explored the potential of DL in omics-based training for prediction of tumor response to therapy, monitoring tumor response during treatment, and patient prognosis. Various bioinformatics tools have been developed and applied in the analysis of omics and inter-omics data in cancer. These approaches may improve diagnosis of breast cancer[38], enhance classification of cancer[39], identify cancer-associated sub-pathways[40], and provide insight into the oncogenic mechanisms and molecular biomarkers in malignant gliomas[41]. Further application of ML/DL in omics datasets in combination with other input data such as radiological images and digitized histopathology has been shown to enhance the power of AI for precision oncology.
MULTI-MODAL ML FOR PRECISION ONCOLOGY
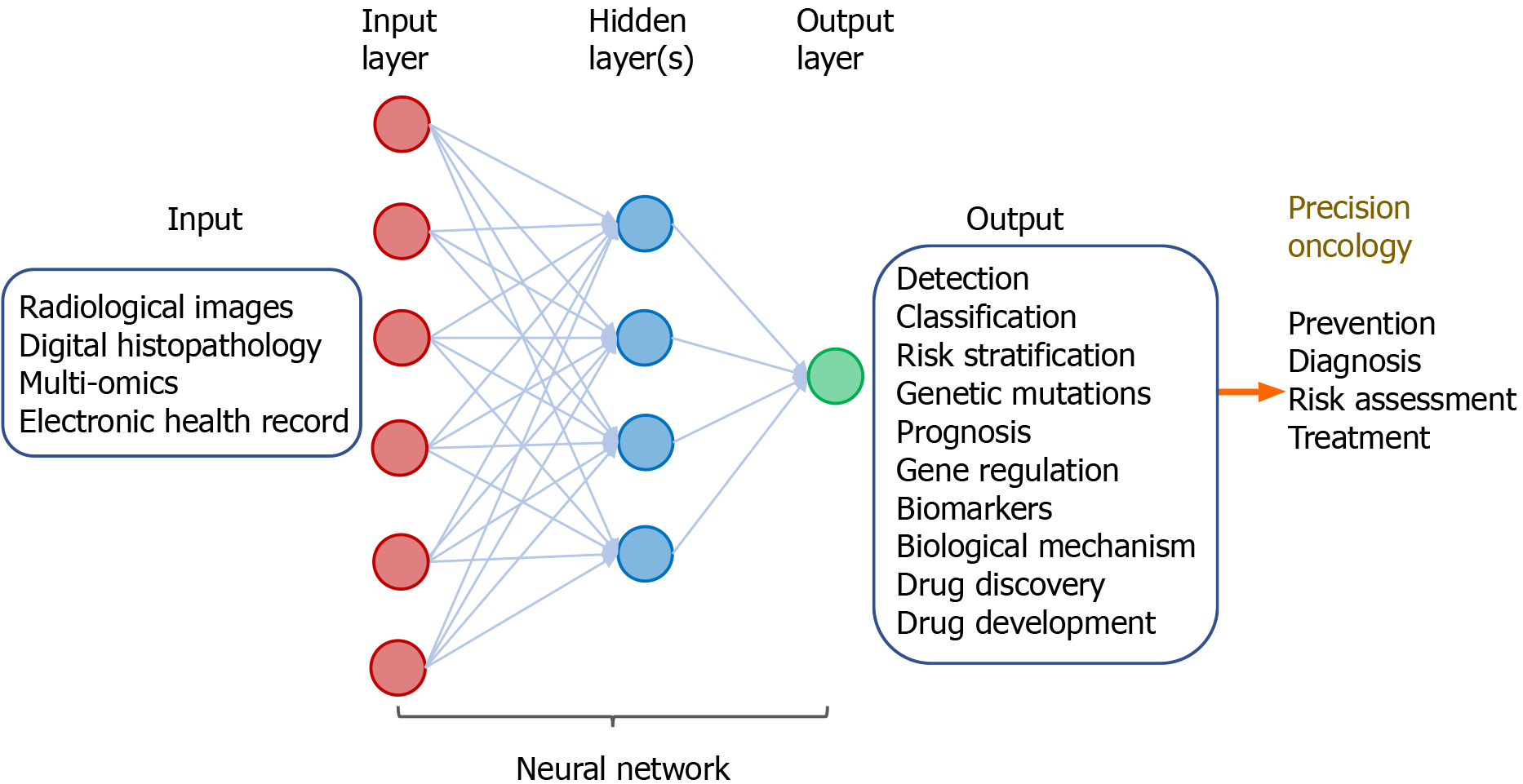
ML by integrative analysis of large data pools combining different types of inputs has been demonstrated to improve accuracy for prediction of diagnosis and clinical outcomes. This involves multi-modal ML in combination of radiological images, digitized histopathology, and omics in conjunction with electronic clinical data. These algorithm-based network models help support and facilitate clinical decision on diagnosis, prognostication, treatment, and patient stratification (Figure 6).
Figure 6 Multi-modal deep learning for precision oncology.
The input comprises clinicopathological datasets from healthy individuals and patients with cancers. The output includes a variety of predicted outcomes that form the basis of precision oncology.
The power of multi-modal ML for precision oncology has been demonstrated by a “holomics” approach that combines medical images, histology, multi-omics, and clinical parameters[42]. By associating radiographic features with patterns of gene expression, a method known as “radiogenomics”, the algorithm-based data produce information on the underlying disease processes and enable prediction of molecular subtypes. Radiomic features can also been linked with other omics data such as proteomics, metabolomics, and immunomics. The radiomic biomarkers being generated can serve as surrogates that help facilitate diagnosis, prognostication, and prediction of tumor response to treatment[43]. In a recent article, a large number of studies in radiogenomics across a variety of tumor sites was reviewed[44]. These include tumors in the brain, lung, breast, ovary, liver, colon/rectum, prostate, and kidney. In these radiogenomics studies, the imaging features of the tumors can be linked with specific cancer genetic mutations. Results of these studies suggest imaging signatures can be developed as predictive biomarkers of genetic alterations in the tumor.
Besides radiogenomics, multi-modal ML of other types of input data have been combined to generate prediction models of clinical outcomes. A computational approach using DL-based CNN combines learning of digitized images of histopathology and genomic biomarkers along with clinical data of patients with glioma. This inter-modal ML-based algorithm has led to development of a predictive model for determining patient survival[45]. By combined analysis of radiomic features of prostate gland with radiologist’s evaluation, prostate specific antigen density, and digital rectal examination, models were developed to characterize prostatic lesions as benign, clinically significant or insignificant cancer. The ML-based models help facilitate selection of patients for MRI-guided biopsy for detection of prostate tumor[46].
Multi-modal ML approaches in combining radiomics and genomics as well as other omics have held great promise for improving capability and accuracy of prediction models in clinical oncology (Figure 6). Application of multi-modal ML for various aspects of translational cancer research is expected to continue to expand. The technical and personnel limitations of this evolving field will need to tackled and resolved. Standardization of ML-based tools along with concerted efforts through collaboration among clinicians and information technologists will help accelerate implementation of multi-modal ML in diagnosis and treatment of cancer.
CONCLUSION
Medical application of AI technology has been revolutionizing healthcare. ML and DL algorithms create powerful tools and opportunities for advancing translational cancer research. Accumulating evidence has begun to demonstrate the value for improving various aspects of clinical oncology such as diagnosis and treatment of cancer. In particular, advances have been made by DNN-based analysis of “big cancer data” towards the goal of precision oncology (Figure 7).
Figure 7 Machine intelligence in translational cancer research for precision oncology.
A number of hurdles will need to be resolved in order to move toward implementation of multi-modal ML in clinical practice. Limitations in radiomics may include inter-observer variability of data processing, reproducibility of radiomic features, tumor heterogeneity, and difference in radiomics approach among researchers[8,44]. Large infrastructural networks and platforms for collection, processing, storage, sharing, and accessing medical images, histopathology, and clinical data across institutions may impose challenges[47]. Due to access of personal information and cloud-based storage of data, ethical and regulatory issues concerning patient confidentiality and data security are non-trivial[48-50].
However, multi-modal ML approaches that integrate large datasets, including medical images, digitalized pathology, holomics, and clinical features will continue to evolve. Emerging applications of AI in oncology involve ML in selection of treatment[23], palliative care and hospice[51], and design of clinical trials[52]. Multi-disciplinary collaboration for development and adoption of multi-modal ML is expected to accelerate healthcare evolution towards precision oncology through computer-aided clinical decision on individualized management of patients.
Manuscript source: Invited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tran B, Tsui SKW S-Editor: Gao CC L-Editor: A P-Editor: Wang LL