INTRODUCTION
As modern science continuously spawns even more dramatic discoveries about human health, more research with the help of new technology and advanced laboratories is increasingly encouraged to find the practical application that can improve human lives. The problems encountered in clinical situation undergo laboratory studies through the problem-solving process (such as finding etiology and pathogenesis to find a key factor against the development of the cause or drugs) and then return to clinical guided treatment, and this process is summarized as from the bedside to the laboratory and vice-versa (Bench to Bedside and Bedside to Bench, i.e., B to B to B, defined as Translational Medicine). Furthermore there are different processes in translational medicine, with the main objective being translation journey from bench to bedside to community. As the medicine enters the era of translational medicine, medical mycology as an important part of medicine has become translational medical mycology[1,2].
Although in translational medical mycology the patient’s history and clinical manifestations are important clues to clinical diagnosis, sample collection from lesion sites is a crucial step in order to identify patients with fungal infection specimens (scales, pus, sputum, blood, cerebrospinal fluid, etc.). Obtaining high quality specimens to meet the laboratory requirements for etiological diagnosis is critical, including professional training in mycology, biologically secure laboratory conditions, preparation of different media, culture incubator (24 °C-26 °C and 37 °C), and most importantly avoidance of contamination during the procedure. Microscopic computer image acquisition and processing system is the basic premise of fungal microscopic examination, specimens are processed (KOH preparation) to observe fungi (hyphae/spores, different shapes and size), and for further inquiry there is also molecular identification. It is estimated that there are more than 1.5 million kinds of fungi on the Earth. So far more than 100000 kinds have been described, but only just over 100 kinds are known as common pathogenic fungi, suggesting that there is a great development space for medical mycology.
The rapid developed molecular biology techniques are used for molecular identification of not only fungi isolated in culture but also those from non-culture specimens, like direct DNA extraction using universal fungal primers by polymerase chain reaction (PCR) amplification/sequencing; further pathological examination (Periodic acid-Schiff or Methenamine-silver staining) can be used to identify the fungus and its special structures. Electron microscopy is used to observe the ultra-structure and fungal pathogens for species identification and study of pathogenesis. For translational medical mycology, molecular biology and individualized treatment should include both the host and the fungus. To perform host susceptibility analysis to the pathogenic species and the virulence factors, molecular biology techniques are required to analyze the host susceptibility genes, environmental and exposure factors. DNA from the fungal specimens (isolated colonies in culture, non-cultured clinical specimens or pathological paraffin block specimens) for molecular amplification and sequencing of pathogenic fungi is equally important. For identification, common fungal universal primers to amplify the internal transcribed spacer 1/4 region and D1/D2 domain are used, but species-specific primers can also be designed.
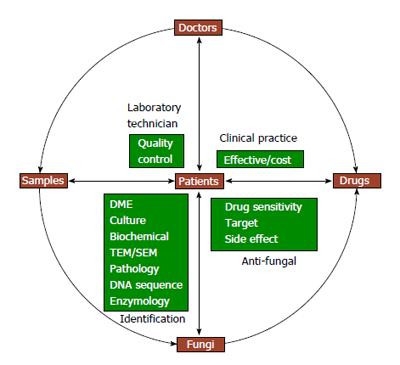
Various comprehensive detection techniques are designed to quickly identify pathogenic fungi for administration of antifungal therapy on time. Antifungal drug sensitivity test is a superior choice to select drugs to which the particular strain is highly sensitive, but at the same time, drug interactions, safety and drug prices should be taken into consideration. Each link has a puzzle challenge with a variety of problems and each clinical problem is transformed into medical mycology research topic. The essential factors for translational medical mycology and their relationship are summarized in Figure 1.
Figure 1 Essential factors for translational medical mycology.
DME: Direct microscopic examination; TEM: Transmission electron microscopy; SEM: Scanning electron microscopy.
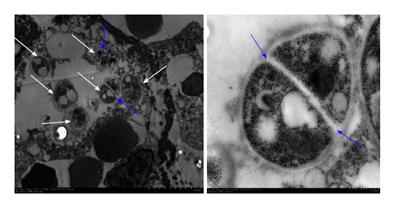
With the improvement in patient acceptance and the rate of positive fungal results[3], we can improve the traditional methods for sample collection. In one patient with Candidosis intertrigo in the groin, specimens were collected using a cotton swab, tape and blunt scraping of scales, respectively, for direct DNA extraction by PCR/sequencing. The results confirmed that the three different methods all identified the pathogenic fungi as Candida albicans, and the results were verified by cultured colony identification. The new method for sample collection by using a cotton swab, tape and using blunt scraping of scales for direct DNA extraction by PCR/sequencing does not only solve the problem of sample collection from sensitive parts, but also can be used for early identification of pathologic fungi and treatment accordingly, which in turn saves time[4]. Direct DNA extraction from the clinical samples (hair or nail) for identification of the pathogenic strains has also been successfully developed to distinguish between tinea capitis[5] or onychomycosis. In one case, hair samples collected from a 6-year-old girl who suffered from kerion were divided into two parts for culture dependent and independent methods, respectively. Since the patient was already on terbinafine, the culture grew slowly. Two sequences from the hair and the culture were both identified as Arthroderma vanbreuseghemii. Hence, PCR based molecular biological validation of DNA directly from the sample can rapidly identify the pathogenic fungus, especially after the use of antifungal drugs. Pathological examination is important in the field of medical mycology, but it has its own limitations. It is important to verify the results since specific fungal strains cannot be determined by pathological reports. In a patient with oral ulcers, histopathology reported a diagnosis of “Leishmaniasis”, but the review of the pathological section found that “Leishmania” is more like Penicillium marneffei yeast cells. The oral specimens were negative for fungal culture. To rule out the “Leishmaniasis” and to establish a new diagnosis as “Penicilliosis marneffei”, paraffin-embedded tissue was used to extract DNA using universal primers for PCR, and the obtained sequence was compared with the sequences in database by Blast and found to share a 99% similarity to the Penicillium marneffei. In addition, the paraffin-embedded tissue was prepared for transmission electron microscopy observation, which showed the septated wall, the specific structure for Penicillium marneffei yeast in tissue (Figure 2). The diagnosis was confirmed as a case of HIV-negative primary, localized, penicilliosis marneffei by the non-culture method[6].
Figure 2 In transmission electron microscopic examination, there are 6 organisms with cell wall (white arrows), of which 2 have septated wall (blue arrows) within a macrophage.
This specific structure of Penicillium marneffei helped to clarify the pathogen as Penicillium marneffei.
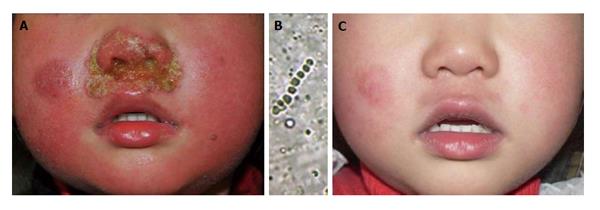
The fungal culture result can be influenced by various factors like culture media, incubation conditions, the volume and quality of inoculation, previous history of use of antifungal drugs and the technique of the inoculation, which lead to unsuccessful attempt to isolate the pathogenic fungi. Direct extraction of DNA is a useful method for guiding treatment in these cases where microscopic tests are positive and cultures are negative. We have encountered some cases in which there was no growth in fungal culture but a significant treatment effect was achieved with antifungal agents[7]. A 3-year-old girl was consulted with impetigo-like lesions around the nostrils. KOH examination showed hyphae and clustered spores in the crust but the culture was negative. The pathogenic agent was identified as Arthroderma vanbreuseghemii, a teleomorph of Trichophyton interdigitale using PCR-based sequencing of the crusts taken from the lesion. She recovered from the infection after oral and topical antifungal agents (Figure 3).
Figure 3 A 3-year-old girl was consulted with impetigo-like lesions around the nostrils.
A: Pustules and erythema around nostrils; B: Hyphae and clustered spores in the crust but the culture was negative. The pathogenic agent was identified as Arthroderma vanbreuseghemii (a teleomorph of Trichophyton interdigitale) using polymerase chain reaction-based sequencing of the crusts; C: The pustules and erythema disappeared after 35 d of antifungal treatment.
Regarding the treatment, whenever the direct microscopic examination with KOH is positive, the antifungal treatment should be started immediately without waiting for the fungus to be isolated or the species be identified. As this might take weeks and the patient’s condition might deteriorate. More importantly, sample collection from the lesions for culture and pathology should be conducted just before starting the antifungal agents. As the treatment progresses, a fungal culture (the conventional method and subsequent identification and susceptibility testing) and direct extraction of DNA for molecular identification should be performed in addition to the weekly follow-up. Samples should be collected and examined until the culture becomes negative before stopping the antifungal treatment[8].
Antifungal drugs are the key factor in the treatment of fungal infection. Commonly used topical antifungal drugs consist of allylamines (such as terbinafine, naftifine, butenafine), imidazoles (such as ketoconazole and bifonazole) or a mixture of the two (naftifine with ketoconazole), and some are even mixed with steroids (triamcinolone acetonide with econazole or miconazole nitrate). Topical drugs are made by mixing various concentrations of antifungal agents with matrices composed of different vehicles and formula. The antifungal spectrum and activity of this mixture (final product) are determined by applying a fixed volume directly to the pathogenic fungus, and then observing and comparing the size of the inhibition zone around the well containing the tested cream. This modified agar diffusion method is a practical agar-based method, which enables determination of the activity of various antifungal drugs against an array of fungal genera and species. It is useful to assess antifungal activity and spectrum against fungal skin infection and to determine the in vitro spectrum and activity of different topical antifungal creams by studying their direct inhibition activity against clinical isolates. We can select the product with the best effect/cost potency ratio from the commercial antifungal creams based on the comparison of the inhibition zone in the same condition to the same species of pathogenic fungus[9,10].
Sometimes in clinical practice we might encounter multiple mixed fungal infections, which will require different media. The samples should be incubated at different temperatures to distinguish them and we must not jump into a premature conclusion that there is a “contaminated fungus”. Generally, the fungus which invades the skin during trauma at the initial stage is considered contaminated. When the local condition becomes suitable for its growth, this “contaminated fungus” could become as the pathogenic fungus (opportunistic fungal infection). This idea leads us to find out the uncommon pathogen and the cure to the difficult cases[11,12].
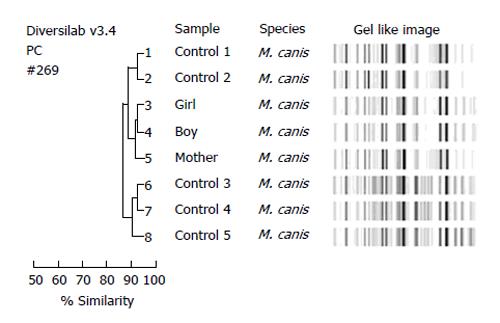
It is important to use techniques of molecular biology and electron microscopy to identify fungi from all the lesions, particularly in cases of rare fungi, where there is not much of a morphological characteristic. In one case, several samples were collected from patient’s face, neck, back, buttocks, foot as well as toenail lesions and all were isolated as Trichophyton rubrum. By using Trichophyton rubrum specific random amplified polymorphic DNA (RAPD) and tandem repeat subunit Trs-1/Trs-2 PCR product analysis, we found that the patient was infected with six different strains at various sites of the infection, proving that various strains did not occur by autoinoculation[13]. RAPD and Trs-1/Trs-2 analysis of 8 family members showed that although Trichophyton rubrum can infect one member of the family to another (2 families), but more cases showed that the infections are beyond the families[14]. The members from one family had kerion (son) and tinea corporis (father and mother), and Arthroderma vanbreuseghemii was isolated from their lesions. By using the morphology, rapid urease test, hair perforation experiment, random primers polymorphism and zymogram analysis, it was proved that the pathogen was the same and it was transmitted from the neighbor’s rabbit to the son and later to his parents[15]. In another case, two children (tinea capitis) and mother (tinea corporis) were diagnosed based on the positive KOH examination. Morphological characteristics and sequencing of the internal transcribed spacers 1 and 2 amplified from primary culture isolates confirmed that their infections were caused by the zoophilic Microsporum canis. Repetitive sequence-based molecular typing using the DiversiLab system, secreted enzymatic activity analysis and antifungal susceptibility indicated that these isolates might share the same source (Figure 4)[16]. Therefore, molecular identification can give us clues about the source of infection whether the infection is caused by autoinoculation, by some external sources or by close contact within the family members.
Figure 4 Molecular typing of eight Microsporum canis isolates showed distinct and similar fingerprint patterns visualized by the gel-like image and as indicated by a > 90% similarity coefficient in the dendrogram.
The three isolates from patients of a same family (number 3, 4, 5) were grouped together with a ≥ 98% similarity compared with the control isolates of M. canis. M. canis: Microsporum canis.
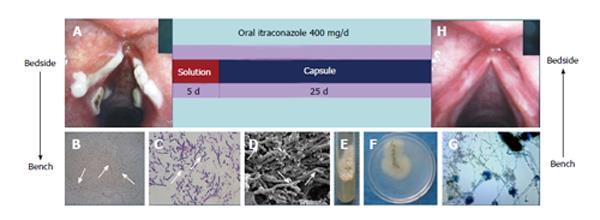
In recent years we have seen several cases of common fungi causing debilitating rare diseases, which give us a new thought about how people with normal immune system can get infected with common pathogens. In one case, a boy had scalp laceration from a road accident, and the treatment included debridement and scalp transplantation in orthopedic surgery. After the treatment, the scalp had honeycomb nodules and abscesses. Culture, morphology and molecular identification showed that it was Microsporum gypseum (M. gypseum) infection. M. gypseum is a geophilic fungus commonly found in soil, and it may have lurked in the scalp when the wound came in contact with soil during the accident and caused secondary infection after incomplete debridement[17]. A patient with severe facial seborrheic dermatitis had a fever of unknown origin. The direct microscopic examination of his face scales showed a large number of Malassezia yeast cells. Therefore, we treated him with itraconazole, and one day after the administration of antifungal drug his temperature dropped down and became normal, and seborrheic dermatitis was significantly improved during three weeks of antifungal therapy. This gives us a picture that systemic fungal infection can be the cause of the fever, though routine fungal culture of blood and even cerebrospinal fluid was negative. Since Malassezia is a lipophilic fungus that needs oil in the medium for the growth, but usually the general laboratory does not provide this type of medium[18]. A woman with severe hoarseness in whom antibiotic treatment did not improve the symptoms received fibrolaryngoscopic examination, and it was discovered that there were plaques on the vocal cord. By using microscopic examination, culture and molecular identification, the results showed Aspergillus fumigatus (A. fumigatus) and later she was cured with itraconazole[19]. In another a woman who had hoarseness because of a lump on her vocal cord, examination of tissues extracted for scanning electron microscopy revealed that they were destroyed by the hyphae of A. fumigatus. In addition, we also discovered several kinds of exocrine protease activities in the specimen[20]. The third female patient with a similar pathogen had severe hoarseness and a medical history of oral sex. The pathogen was identified by morphology and molecular identification. Treatment was successful by oral itraconazole solution for the first five days and then it was followed by capsules, which completely cured the patient in 30 d (Figure 5). This case gives us a new thought about how people with normal immune system can be infected with common airborne pathogenic A. fumigates. The vocal mucous membrane barrier which was damaged by oral sex made it easy for the colonization of the spore which later invades the tissue[21].
Figure 5 Treatment was successful with oral itraconazole solution the first five days followed by capsules, which completely cured the patient in 30 d.
A: A laryngoscopy was performed and revealed obvious white plaques on the swollen vocal cords and laryngeal ventricle. Samples were taken from the vocal cords; B: Numerous hyphae branching at 45° angles were detected by light microscopy; C: Pathology; D: Scanning electron microscopy; E and F: A velvety and powdery colony developed on Sabouraud Dextrose Agar at 28 °C; G: Microscopic examination of a slide culture revealed the morphologic features consistent with those of Aspergillus fumigatus. The patient was treated with oral itraconazole solution followed by capsules for 30 d; H: Laryngoscopy showed that the vocal cords were smooth without any white plaques.
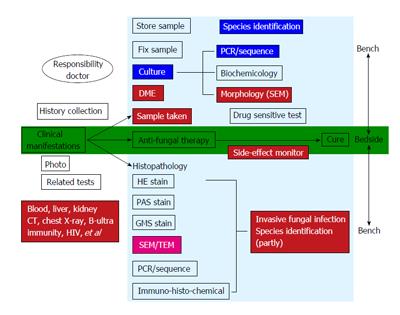
Over the past one century medical mycology has had a typical “transformation” characteristic: we used to take samples from the lesion of a patient to check the fungal elements under the microscope to make the diagnosis and then treat the patient. Translational Medical Mycology is not just an empty theory and slogan, it should be in the classic “transformation” based on constantly enhanced (electron microscopy, etc.) and enriched contents (molecular biology, etc.) and individual medicine. In summary, in “translational medicine mycology” the starting point and end point are the clinical and laboratory studies, which are aimed to solve clinical problems (Figure 6). Patient is the “center” and problems that are encountered during the diagnosis and treatment are transforming medical mycology. For specific diseases, we might need to integrate microbiologists, pathologists, molecular biologists and other laboratory experts with all techniques and methods to determine the pathogenic fungi, help with susceptibility testing, remote consultation, so that we can start the treatment as soon as possible to save patient lives. Transformation is a dynamic, multi-level, multi-directional and continuously improving process. We can solve a clinical problem by using a number of new technologies, new methods and training, and then there will be an improvement in the treatment of fungal diseases. It will eventually promote the overall progress of medical mycology and ultimately provide an access to the effective treatment of the disease as well as prevention.
Figure 6 Translational medical mycology diagnosis and treatment pathway-from bedside to bench to bedside.
CT: Computed tomography; DME: Direct microscopic examination; TEM: Transmission electron microscopy; SEM: Scanning electron microscopy; HE stain: Haematoxylin and eosin stain; GMS stain: Gomort methenamine silver stain; PAS stain: Periodic Acid Schiff stain; PCR: Polymerase chain reaction.