©2013 Baishideng Publishing Group Co.
World J Transl Med. Dec 12, 2013; 2(3): 67-74
Published online Dec 12, 2013. doi: 10.5528/wjtm.v2.i3.67
Published online Dec 12, 2013. doi: 10.5528/wjtm.v2.i3.67
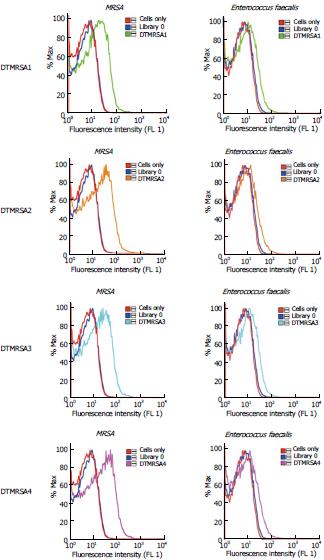
Figure 1 Flow cytometry histograms of the relevant aptamer candidates.
Four aptamers show significant binding to methicillin-resistant Staphylococcus aureus (MRSA) 43300 bacteria, but not to the Enterococcus faecalis cell line.
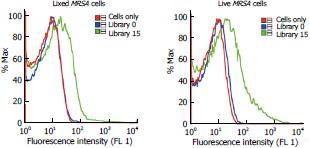
Figure 2 Comparison of the binding results between fixed and live methicillin-resistant Staphylococcus aureus cells.
Binding assays show that results remain the same, irrespective of whether experiments are run with fixed or live cells. MRSA:Methicillin-resistant Staphylococcus aureus.
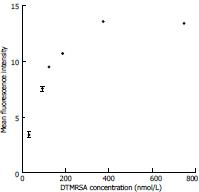
Figure 3 SigmaPlot binding curve of the aptamer DTMRSA4 used for the Kd value determination.
Methicillin-resistant Staphylococcus aureus cells were incubated with various concentrations of fluorescein isothiocyanate-labeled DTMRSA4.
- Citation: Turek D, Simaeys DV, Johnson J, Ocsoy I, Tan W. Molecular recognition of live methicillin-resistant staphylococcus aureus cells using DNA aptamers. World J Transl Med 2013; 2(3): 67-74
- URL: https://www.wjgnet.com/2220-6132/full/v2/i3/67.htm
- DOI: https://dx.doi.org/10.5528/wjtm.v2.i3.67