Published online Jun 20, 2020. doi: 10.5496/wjmg.v9.i1.1
Peer-review started: March 14, 2020
First decision: April 12, 2020
Revised: April 19, 2020
Accepted: May 14, 2020
Article in press: May 14, 2020
Published online: June 20, 2020
Processing time: 94 Days and 14.5 Hours
Wiedemann-Steiner syndrome (OMIM #605130) is a rare congenital malformation syndrome characterized by hypertrichosis cubiti associated with short stature; consistent facial features, including long eyelashes, thick or arched eyebrows with a lateral flare, wide nasal bridge, and downslanting and vertically narrow palpebral fissures; mild to moderate intellectual disability; behavioral difficulties; and hypertrichosis on the back. It is caused by heterozygous pathogenic variants in KMT2A. This gene has an established role in histone methylation, which explains the overlap of Wiedemann-Steiner syndrome with other chromatinopathies, a heterogeneous group of syndromic conditions that share a common trigger: The disruption of one of the genes involved in chromatin modification, leading to dysfunction of the epigenetic machinery.
Core tip: Chromatinopathies are a highly heterogeneous group of syndromic conditions in which the underlying genetic anomaly consists of disruption of one of the components of the epigenetic machinery. Within this group, which contains more than 40 diseases, including Kabuki, Sotos, Kleefstra, Koolen-De-Vries/KANSL1 haploinsufficiency, Rubinstein-Taybi, KAT6B-related syndromes, Smith-Magenis, Rett, Townes-Brock, Bohring-Opitz, ATRX, CHARGE, and Floating-Harbor syndromes, an emerging member is represented by Wiedemann-Steiner syndrome, which has very interesting features.
- Citation: Fontana P, Passaretti FF, Maioli M, Cantalupo G, Scarano F, Lonardo F. Clinical and molecular spectrum of Wiedemann-Steiner syndrome, an emerging member of the chromatinopathy family. World J Med Genet 2020; 9(1): 1-11
- URL: https://www.wjgnet.com/2220-3184/full/v9/i1/1.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v9.i1.1
Eukaryotic DNA is organized in a finely packaged structure called chromatin. The degree of the opening of the chromatin varies all along the chromosomes and it can essentially be resumed into two different modes: “Open” chromatin, named euchromatin, which is lightly packed, usually enriched in genes, and accessible to mRNA transcription machinery; and “closed” chromatin, named heterochromatin, which is tightly packed and inaccessible to polymerases. Regions of DNA coding for genes can be transiently condensed and transcriptionally silenced but can switch this state of “facultative heterochromatin” to a euchromatic, transcriptionally active structure[1].
The regulation of the structure of chromatin usually takes place through post-translational modifications, mainly methylation and acetylation, of the core histone proteins, whose binding to DNA, tight because of the histone positive charges, relaxes after the addition of negatively charged groups such as methyl and acetyl groups[2].
The huge number of proteins involved in the modification of chromatin is classified into four main groups of factors: Writers, erasers, readers, and remodelers, with different functions. Proteins methylating or acetylating histones are called writers because they add acetyl or methyl groups to histone proteins. The effect of histone acetylase action is decondensation of a region of chromatin, allowing, subsequentially, the transcription of the corresponding gene. Methylation can occur on lysine and arginine residues only and this post-translational modification is characterized by high specificity since only specific lysines and arginines are involved. Histone methylation can have an activating or a repressive effect on transcription depending on the histone residue that is methylated (methylation of H3K4, H3K36, and H3K79 usually activates transcription, while methylation of H3K9, H3K27, and H4K20 has a repressive effect) and on its position (methylation at H3K36 has a negative effect when it falls in the promoter and not in the coding region)[1].
On the other side, histone deacetylases and histone demethylases act as erasers because they remove acetyl and methyl marks, respectively. Histone deacetylases generally make genes transcriptionally inactive while histone demethylases can make genes active or inactive depending on the residue that is modified. Readers recognize specific post-translational marks on histones and determine their functional effects. Finally, remodelers regulate the balance of chromatin through histone post-translational modifications and modifications in the histone-DNA interaction, which leads to nucleosome sliding or changes in the conformation of nucleosomal DNA[3,4].
The term “chromatinopathies” has been recently adopted, referring to a heterogeneous group of syndromic conditions that share a common trigger: The disruption of one of the genes involved in chromatin modification, leading to dysfunction of the epigenetic machinery. In consideration of the high heterogeneity of genes involved and, above all, the high heterogeneity of the target genes, whose transcription is altered, the clinical features of chromatinopathies are highly variable. Some of them are characterized by short stature and microcephaly, others by overgrowth, and several conditions present with typical patterns of dysmorphic features. The prevalence and type of congenital defects differ greatly from one syndrome to another[5-8].
On the other hand, developmental impairment is almost a constant feature of chromatinopathies. Several studies have highlighted that correct modulation of histone methylation is crucial during neurogenesis and plays a key role in the consolidation of long-term memory and contextual learning[9,10].
Mutations in histone lysine methylation-related genes have been identified in several patients with both syndromic and nonsyndromic intellectual disability[10].
The name Wiedemann-Steiner syndrome (WDSTS) was coined by Koenig et al[11] in 2010, referring to a clinical spectrum recalling two different patients reported by Wiedemann and Steiner, in 1989 and 2000, respectively[12,13]. Wiedemann described a male patient with pre- and post-natal growth deficiency, psychomotor delay, round and flat face, short nose, hypertelorism, long philtrum, short palpebral fissures, low-set ears, and high-arched palate. Steiner reported an 8-year-old girl with short stature, thick eyebrows, telecanthus, broad nasal bridge, long philtrum, thin upper lip, clinodactyly of the fifth finger, mild to moderate psychomotor delay, and hypertrichosis, which became more accentuated with age and was most evident over the limbs, especially at the elbows, and the back.
Hairy elbows in association with short stature and/or developmental delay had been reported, in fact, also by other authors, at least some of them describing, presumably, patients with WDSTS[14-19].
In their paper, Koenig et al[11] collected three cases with developmental delay and a recognizable pattern of phenotypic traits including narrow and downslanting palpebral fissures, hypertelorism, thick eyebrows with synophrys, broad nasal bridge, high-arched palate, clinodactyly, and/or brachydactyly. Two of the three patients also had hypertrichosis at the limbs. These features constitute a typical, recognizable phenotype, distinctive of WDSTS.
In 2012, Jones et al[20] identified the haploinsufficiency of the gene KMT2A (MLL) as the genetic cause of WDSTS. Whole-exome sequencing was performed in six patients with a suggestive phenotype (hypertrichosis cubiti, short stature, intellectual disability, and facial features consistent with the patients reported by Wiedemann and Steiner) and detected de novo loss-of-function mutations in five of the six patients.
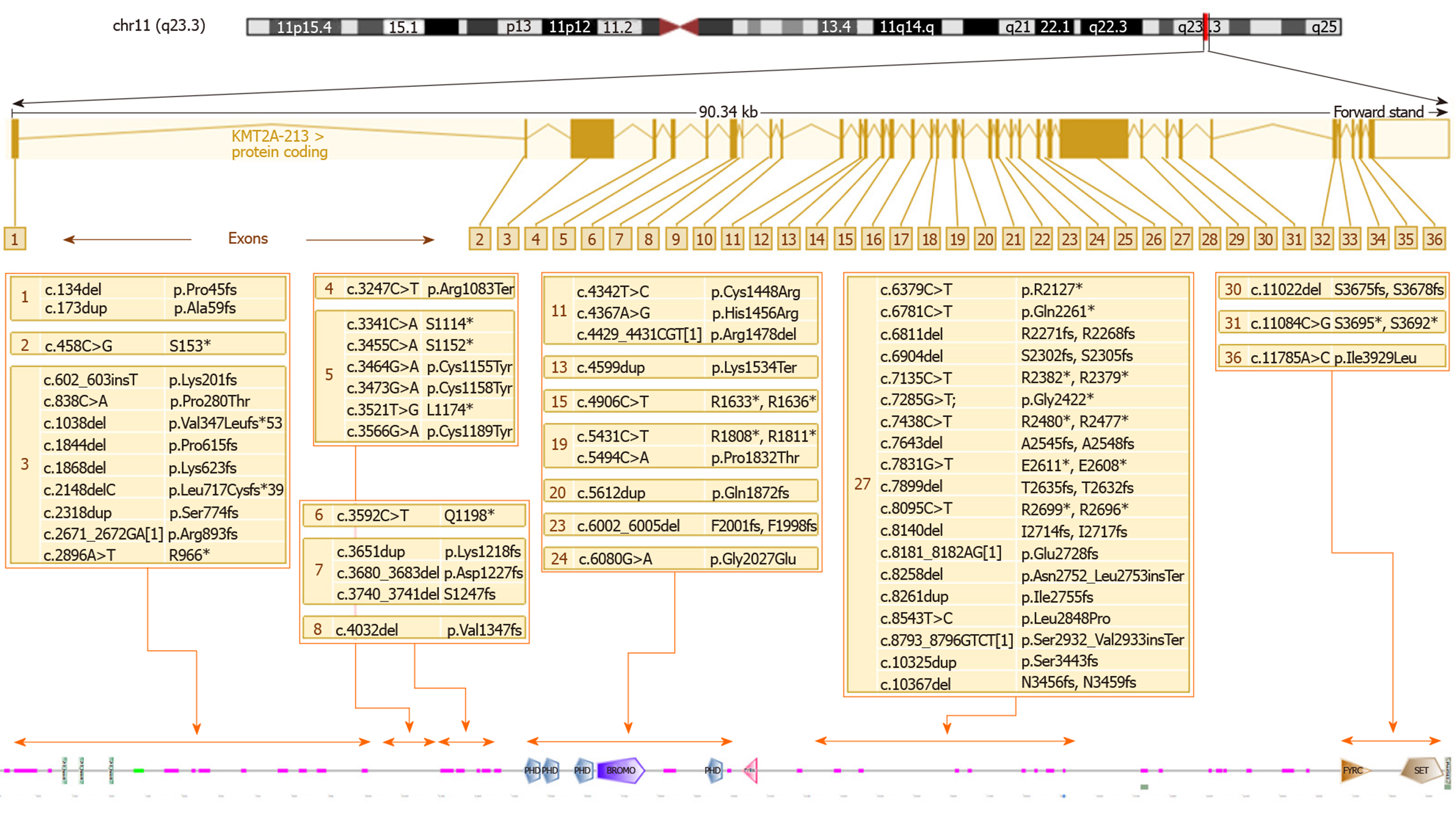
KMT2A encodes a DNA-binding protein that methylates a lysine residue on histone H3 (H3K4). It consists of 37 exons, but a major transcript of 14982 bp produces a 3969 amino acids protein from 36 of the 37 exons. The protein contains several functional domains, including the SET domain, responsible for its H3K4 methyltransferase activity[21]. As discussed above, histone methyltransferases act as “writers”. KMT2A, indeed, positively regulates the expression of many target genes, including genes belonging to the HOX complex and other genes involved in embryonic development[22-24]. Studies on mice also demonstrated that KMT2A is highly expressed in adult hippocampal neurons and is critical for synaptic plasticity, cognition, complex behaviors, and long-term memory[25,26].
Other members of the family of H3K4 methyltransferases are associated with other chromatinopathies, e.g., KMT2D and KMT2B are associated with Kabuki syndrome and Kleefstra syndrome, respectively [10].
As for the other chromatinopathies, the involvement of a transcription factor that modulates the expression of so many genes determines a high heterogeneity of clinical signs, whose frequency has been progressively delineated in the years.
The phenotype includes many characteristic facial features, most of them shared with other chromatinopathies, which overall allow a “gestaltic” diagnosis.
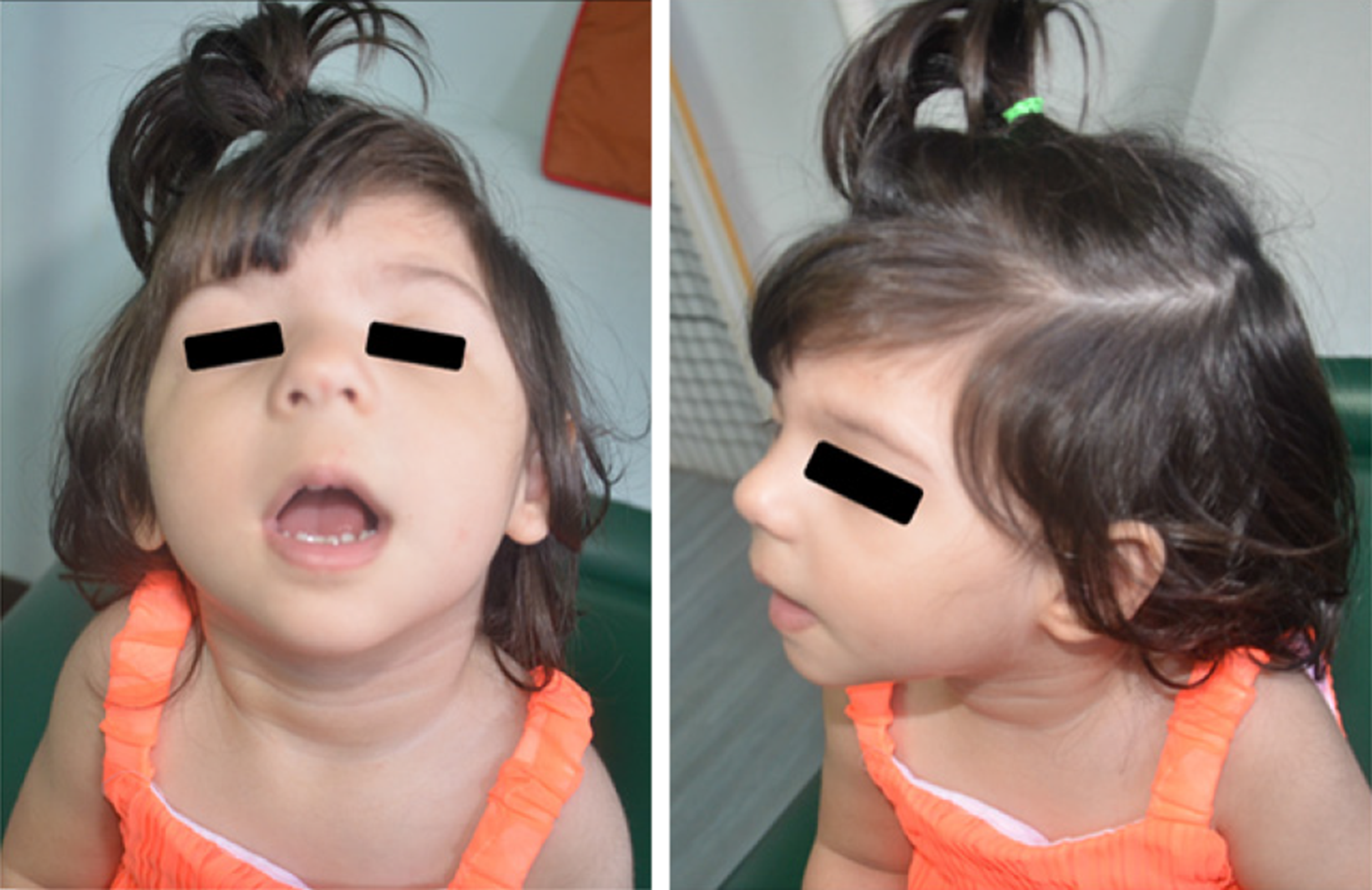
The appearance of the periorbital region is peculiar and characterized by: Broad, arched eyebrows, reported in about 80% of the patients and often associated with synophrys, recalling Cornelia de Lange syndrome; hypertelorism (66%-93%, depending on the cohort); long eyelashes (over 80%); and long and downslanting palpebral fissures (58%-92%) in some way recalling Kabuki syndrome, but often with a marked vertical narrowing which is unusual in the other syndromes of the same family, except for Pierpont syndrome. The nasal bridge is usually wide (71%-89%), with a broad, bulbous tip. Patients often also show low-set ears, long philtrum, thin vermilion, micrognathia, high arched palate, and anomalies of the dentition[21,27-32]. Figure 1 shows a patient with WDSTS who has come to our observation.
As in other chromatinopathies such as Coffin-Siris syndrome, Nicolaides-Baraitser syndrome, Cornelia de Lange syndrome, and Kabuki syndrome, some minor skeletal anomalies are common[33,34]. Clinodactyly of the fifth finger has been seen in more than half of the patients; hands and feet are usually small and puffy, with mild brachydactyly. Congenital hip dislocation has been reported by several authors[27,35,36], while polydactyly is rare[37]. A marked hypertrichosis cubiti (so-called “hairy elbow”) is considered the pathognomonic sign of WDSTS. In the first reports, the frequency of this feature was near to 100% because of the obvious selection bias, since patients with developmental delay, typical facial appearance, and hypertrichosis cubiti were selected for the molecular diagnosis of WDSTS. Recent cohort studies estimate a frequency of about 60%; hairy elbows can be associated with hypertrichosis of the back and/or generalized hypertrichosis[38,39].
Prenatal and postnatal growth retardation is common, while microcephaly has been reported in 33%-56% of the subjects[38,40]. Sun et al[27] reported short stature in all the patients described in the literature before their manuscript, while Baer et al[38] suggested that about half of the patients could present a normal stature. Some evidence suggests that short stature is often due to a growth hormone deficiency, secondary or not to structural pituitary anomalies such as hypoplastic anterior pituitary or ectopic posterior pituitary. For these reasons, a role in pituitary development has been hypothesized for the KMT2A gene. A growth hormone test and a pituitary magnetic resonance scan are suggested for patients with a diagnosis of WDSTS and short stature. The effects of long-term treatment with growth hormone in patients with WDSTS have been rarely described, but the first data suggest that the growth response is good and that prompt identification of a growth hormone deficit allows a significant recovery in final stature[41].
Developmental delay/intellectual disability is the rule for the syndrome. The degree is usually mild to moderate. Neonatal hypotonia is present in more than half of the patients, while seizures are rare. The prevalence of autism has been recently estimated as 11.8%, but patients without autism can also show behavioral anomalies such as repetitiveness, emotional dysregulation, ADHD, anxiety, and hetero- and auto-aggressiveness[35,42,43]. Cerebral malformations are very rare and heterogeneous; corpus callosum malformations seem to be the most recurrent ones.
Patients with WDSTS have a significantly increased risk of developing recurrent respiratory and urinary tract infections, secondary to immunological disorders such as panhypogammaglobulinemia, lymphopenia, and poor antibody responses, requiring frequent hospitalizations and immunoglobulin infusions. For this reason, a complete immunologic profile upon diagnosis is warranted. Increased absolute count of eosinophils has been occasionally described. Immunodeficiency is not uncommon in other chromatinopathies, especially Kabuki syndrome[36,44,45].
About one-third of patients present with congenital heart defects; the most common are patent ductus arteriosus, septal defects, and aortic anomalies[27,32,38]; and a good part of these defects do not require surgical correction. On the other side, patients frequently undergo surgery because of ophthalmologic abnormalities, most commonly ptosis, strabismus, and lachrymal duct anomalies. Surgery for ptosis can be scarcely effective when it is associated with a markedly vertical narrowing of the palpebral fissures. Ocular anomalies overall affect more than half of the individuals.
Urogenital anomalies affect about 30% of the patients, but they are markedly heterogeneous; hypoplasia of the kidneys, horseshoe kidney, and cryptorchidism have been described[29,36,43,46]. A female patient has been reported with a unicornuate uterus and a unique left ovary, fallopian tube, and kidney[44].
Since 2012, when Jones et al[20] first linked WDSTS with mutations in the gene KMT2A, the database of mutations has grown to include more than 60 sequence variations.
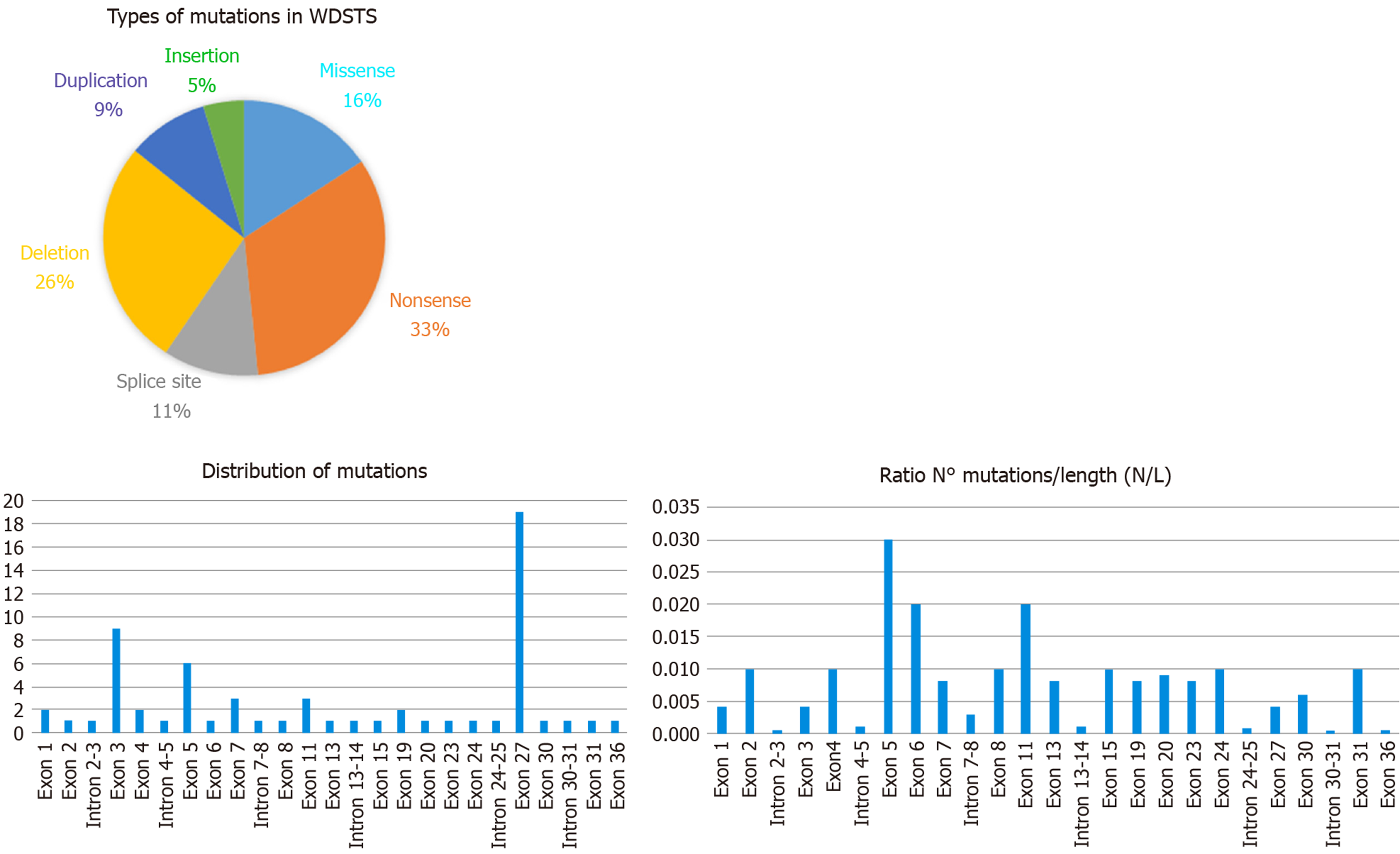
Examining the data from the free versions of the databases ClinVar and HGMD, we have been able to index 64 sequence variations. In detail, they fall into six categories with this respective proportions: Nonsense 33%, deletions 26%, missense 16%, splice site 11%, duplications 9%, and insertions 5% (see Figure 2). All of them are classified according to the ACMG/AMP 2015 guideline as Pathogenic or Likely Pathogenic (as reported in Table 1).
| No. | cDNA | Transcript | Protein change | Gene region | Mutation type | Clinical interpretation | refSNP cluster | Database |
| 1 | c.134del | NM_001197104.1 | p.Pro45fs | Exon 1 | Deletion | Pathogenic | rs1555138529 | ClinVar/HGMD |
| 2 | c.173dup | NM_001197104.2 | p.Ala59fs | Exon 1 | Duplication | Pathogenic | ClinVar | |
| 3 | c.458C>G | NM_001197104.1 | S153* | Exon 2 | Nonsense | Pathogenic | rs587783678 | ClinVar |
| 4 | c.502+1G>A | NM_001197104.2 | Intron 2-3 | Splice site | Pathogenic | ClinVar | ||
| 5 | c.602_603insT | NM_005933.4 | p.Lys201fs | Exon 3 | Insertion | Pathogenic | rs1555035550 | ClinVar |
| 6 | c.838C>A | NM_001197104.1 | p.Pro280Thr | Exon 3 | Missense | Likely Pathogenic | HGMD | |
| 7 | c.1038del | NM_005933.4 | p.Val347Leufs*53 | Exon 3 | Deletion | Pathogenic | rs1555035779 | ClinVar/HGMD |
| 8 | c.1844del | NM_001197104.2 | P615fs | Exon 3 | Deletion | Pathogenic | ClinVar | |
| 9 | c.1868del | NM_001197104.1 | p.Lys623fs | Exon 3 | Deletion | Pathogenic | rs797044937 | HGMD |
| 10 | c.2148delC | NM_001197104.1 | p.Leu717Cysfs*39 | Exon 3 | Deletion | Pathogenic | HGMD | |
| 11 | c.2318dup | NM_005933.4 | p.Ser774fs | Exon 3 | Duplication | Pathogenic | rs782297546 | ClinVar/HGMD |
| 12 | c.2671_2672GA[1] | NM_005933.4 | p.Arg893fs | Exon 3 | Insertion | Pathogenic | rs587783676 | ClinVar |
| 13 | c.2896A>T | NM_001197104.1 | R966* | Exon 3 | Nonsense | Pathogenic | rs1555036801 | ClinVar |
| 14 | c.3157-7_3161del | NM_001197104.2 | 4 (intron 3-4) | Deletion | Pathogenic | ClinVar | ||
| 15 | c.3247C>T | NM_001197104.1 | p.Arg1083Ter | Exon 4 | Nonsense | Pathogenic | rs782451966 | HGMD |
| 16 | c.3334+1G>A | NM_001197104.1 | Intron 4-5 | Splice site | Pathogenic | rs1135401764 | ClinVar | |
| 17 | c.3341C>A | NM_001197104.1 | S1114* | Exon 5 | Nonsense | Pathogenic | rs1555038029 | ClinVar |
| 18 | c.3455C>A | NM_001197104.2 | S1152* | Exon 5 | Nonsense | Pathogenic | ClinVar | |
| 19 | c.3464G>A | NM_001197104.1 | p.Cys1155Tyr | Exon 5 | Missense | Likely Pathogenic | rs1057518074 | HGMD |
| 20 | c.3473G>A | NM_001197104.1 | p.Cys1158Tyr | Exon 5 | Missense | Likely Pathogenic | rs1131691503 | ClinVar |
| 21 | c.3521T>G | NM_001197104.1 | L1174* | Exon 5 | Nonsense | Pathogenic | rs1555038111 | ClinVar |
| 22 | c.3566G>A | NM_005933.4 | p.Cys1189Tyr | Exon 5 | Missense | Pathogenic | rs1555038125 | ClinVar/HGMD |
| 23 | c.3592C>T | NM_001197104.2 | Q1198* | Exon 6 | Nonsense | Pathogenic | ClinVar | |
| 24 | c.3651dup | NM_005933.4 | p.Lys1218fs | Exon 7 | Duplication | Pathogenic | rs863224887 | ClinVar |
| 25 | c.3680_3683del | NM_001197104.2 | p.Asp1227fs | Exon 7 | Deletion | Pathogenic | ClinVar | |
| 26 | c.3740_3741del | NM_001197104.1 | S1247fs | Exon 7 | Deletion | Likely Pathogenic | rs1565286640 | ClinVar |
| 27 | c.4012+2T>A | NM_005933.4 | Intron 7-8 | Splice site | Pathogenic | ClinVar | ||
| 28 | c.4032del | NM_005933.4 | p.Val1347fs | Exon 8 | Deletion | Pathogenic | ClinVar | |
| 29 | c.4086+1G>A | NM_001197104.1 | Intron 8-9 | Splice site | Likely Pathogenic | rs863224889 | ClinVar/HGMD | |
| 30 | c.4342T>C | NM_001197104.1 | p.Cys1448Arg | Exon 11 | Missense | Likely Pathogenic | rs863224895 | ClinVar/HGMD |
| 31 | c.4367A>G | NM_001197104.1 | p.His1456Arg | Exon 11 | Missense | Likely Pathogenic | rs1131691433 | ClinVar |
| 32 | c.4429_4431CGT[1] | NM_001197104.2 | p.Arg1478del | Exon 11 | Nonsense | Likely Pathogenic | ClinVar | |
| 33 | c.4599dup | NM_005933.4 | p.Lys1534Ter | Exon 13 | Nonsense | Pathogenic | rs398122881 | ClinVar/HGMD |
| 34 | c.4696+1G>A | NM_001197104.1 | Intron 13-14 | Splice site | Pathogenic | rs1057519407 | ClinVar | |
| 35 | c.4906C>T | NM_001197104.2 | R1633*, R1636* | Exon 15 | Nonsense | Likely Pathogenic | ClinVar/HGMD | |
| 36 | c.5431C>T | NM_001197104.2 | R1808*, R1811* | Exon 19 | Nonsense | Likely Pathogenic | ClinVar | |
| 37 | c.5494C>A | NM_001197104.1 | P1832T, P1829T | Exon 19 | Missense | Likely Pathogenic | rs797045051 | ClinVar |
| 38 | c.5612dup | NM_005933.4 | p.Gln1872fs | Exon 20 | Duplication | Pathogenic | rs1555043939 | ClinVar |
| 39 | c.6002_6005del | NM_001197104.1 | F2001fs, F1998fs | Exon 23 | Deletion | Pathogenic | rs1057519408 | ClinVar |
| 40 | c.6080G>A | NM_001197104.1 | G2027E, G2024E | Exon 24 | Missense | Likely Pathogenic | rs1057519403 | ClinVar |
| 41 | c.6158+6T>C | NM_001197104.1 | Intron 24-25 | Splice site | Likely Pathogenic | rs1555045177 | ClinVar | |
| 42 | c.6379C>T | NM_001197104.1 | p.R2127* | Exon 27 | Nonsense | Pathogenic | HGMD | |
| 43 | c.6781C>T | NM_001197104.1 | p.Gln2261* | Exon 27 | Nonsense | Pathogenic | HGMD | |
| 44 | c.6811del | NM_001197104.1 | R2271fs, R2268fs | Exon 27 | Deletion | Pathogenic | rs797045656 | ClinVar |
| 45 | c.6904del | NM_005933.4 | S2302fs, S2305fs | Exon 27 | Deletion | Pathogenic | rs398122880 | ClinVar/HGMD |
| 46 | c.7135C>T | NM_005933.4 | R2382*, R2379* | Exon 27 | Nonsense | Pathogenic | rs387907275 | ClinVar/HGMD |
| 47 | c.7285G>T | NM_001197104.1 | p.Gly2422* | Exon 27 | Nonsense | Likely Pathogenic | HGMD | |
| 48 | c.7438C>T | NM_001197104.1 | R2480*, R2477* | Exon 27 | Nonsense | Pathogenic | rs1555046568 | ClinVar/HGMD |
| 49 | c.7643del | NM_001197104.2 | A2545fs, A2548fs | Exon 27 | Deletion | Pathogenic | ClinVar | |
| 50 | c.7831G>T | NM_001197104.1 | E2611*, E2608* | Exon 27 | Nonsense | Pathogenic | rs587783679 | ClinVar |
| 51 | c.7899del | NM_001197104.2 | T2635fs, T2632fs | Exon 27 | Deletion | Pathogenic | ClinVar | |
| 52 | c.8095C>T | NM_001197104.1 | R2699*, R2696* | Exon 27 | Nonsense | Pathogenic | rs587783680 | ClinVar |
| 53 | c.8140del | NM_005933.4 | I2714fs, I2717fs | Exon 27 | Deletion | Pathogenic | rs1131692268 | ClinVar/HGMD |
| 54 | c.8181_8182AG[1] | NM_001197104.2 | p.Glu2728fs | Exon 27 | Insertion | Likely Pathogenic | ClinVar | |
| 55 | c.8258del | NM_005933.4 | p.Asn2752_Leu2753insTer | Exon 27 | Nonsense | Pathogenic | rs398122879 | ClinVar |
| 56 | c.8261dup | NM_005933.4 | p.Ile2755fs | Exon 27 | Duplication | Pathogenic | rs1565304395 | ClinVar |
| 57 | c.8543T>C | NM_001197104.1 | L2848P, L2845P | Exon 27 | Missense | Likely Pathogenic | rs1555047266 | ClinVar |
| 58 | c.8793_8796GTCT[1] | NM_005933.4 | p.Ser2932_Val2933insTer | Exon 27 | Nonsense | Pathogenic | rs398122878 | ClinVar/HGMD |
| 59 | c.10325dup | NM_005933.4 | p.Ser3443fs | Exon 27 | Duplication | Pathogenic | rs863224888 | ClinVar |
| 60 | c.10367del | NM_005933.4 | N3456fs, N3459fs | Exon 27 | Deletion | Pathogenic | ClinVar | |
| 61 | c.11022del | NM_005933.4 | S3675fs, S3678fs | Exon 30 | Deletion | Pathogenic | rs1565310297 | ClinVar |
| 62 | c.11071+1G>A | NM_001197104.1 | Intron 30-31 | Splice site | Pathogenic | rs1555049702 | ClinVar | |
| 63 | c.11084C>G | NM_001197104.1 | S3695*, S3692* | Exon 31 | Nonsense | Pathogenic | rs782477344 | ClinVar |
| 64 | c.11785A>C | NM_001197104.2 | I3926L, I3929L | Exon 36 | Missense | Likely Pathogenic | ClinVar |
KMT2A consists of 36 exons and the variants are located quite uniformly along the sequence of the gene. Several authors have noticed that the greatest number of mutations are in exon 27, consistent with the observation that it is the longest exon[38]. By comparing the number of variants (N) with the length of exon/intron (L), it appears that exon 5 (0.03 ratio N/L) displays the highest density of mutations (as shown in Figure 2), while exon 27, the greatest exon of the gene, has a lower ratio (0.004 ratio N/L).
Figure 3 shows the WDSTS-associated variants, subdivided according to the exon in which they map and grouped according to their position in relation to domains, repeats, motifs, and features of the protein.
Several authors have underlined the complexity of the genotype-phenotype correlation in this syndrome. Baer et al[38] suggest as an example a patient with severe intellectual disability and absence of speech, sharing the same missense mutation with a patient only presenting a mild intellectual disability. We can presume that the complex interactions of the methyltransferase encoded by KMT2A with a multiplicity of target genes and other modulators of transcription can determine the high phenotypic variability, even within single families. Another element worth of consideration is that more than a half of the molecular defects reported are nonsense mutations or large deletions; these alterations usually lead to the synthesis of a truncated protein with no functional effects or the absence of the protein because of mRNA decay and not to the malfunction of a single domain. Thus, haploinsufficiency of KMT2A is the main pathogenic mechanism underlying WDSTS. Among the missense mutations, we can notice a cluster located in the first exons of the gene and disrupting the cysteine-rich zinc finger domain implicated in DNA binding. These mutations, perhaps eliciting a dominant-negative effect, seem to be associated with a more severe neurodevelopmental phenotype, with a more severe intellectual disability and a higher prevalence of hypotonia and seizures[32]. Missense mutations in this domain could also be related to a higher prevalence of immunodeficiency, hypogammaglobulinemia, and recurrent infections[21,30,36]. On the other hand, at least one family with panhypogammaglobulinemia and severe recurrent respiratory infections has been reported having a splice site mutation leading to the skipping of exon 28, markedly distal from the DNA binding domain[44].
As mentioned above, mutations in KMT2A have been detected in patients with clinical diagnoses different from WDSTS, but still belonging to the family of chromatinopathies. On the other side, patients with features reminiscent of WDSTS have been found to have mutations in genes related to other syndromes. Because of the overlap of several clinical features, a large panel of genes related to chromatinopathies should be taken into consideration, when available, as the first option in the diagnosis of individuals with a phenotype consistent with WDSTS. Recently, in a small number of patients with an overlapping phenotype, homozygous point mutations (both miss-sense and non-sense) or deletions of the TASP1 gene have been found. The subjects showed developmental delay, hypotonia, microcephaly, recurrent respiratory infections, cryptorchidism (in males), minor heart and kidney anomalies, hirsutism, minor limb skeletal anomalies, and several suggestive facial features (thick, arched eyebrows with synophrys, hypertelorism, epicanthus, broad nasal bridge, and low-set ears)[47]. Thus, loss-of-function mutations of TASP1 could represent the cause of an autosomal recessive form of WDSTS, for which TASP1 may be considered a second disease-causing gene. However, further evidence is needed to determine if a unique phenotype or two different clinical entities are ascribable to mutations in KMT2A and TASP1. From a biological point of view, these two genes are strictly connected because taspase 1, the enzyme codified by TASP1, is crucial for the cleavage, processing and, consequently, activation of KMT2A. The loss of function of TASP1 is presumed to determine a lack of activation of KMT2A and downregulation of the expression of the downstream genes, including genes belonging to the HOX complex[48]. However, TASP1 also processes other modulators of transcriptions including KMT2D, the gene related to Kabuki syndrome, so the related phenotype is supposed to be highly heterogeneous.
Disorders caused by mutations in genes regulating chromatin remodeling are called chromatinopathies. Chromatinopathies are characterized by peculiar features, both from a clinical and genetic point of view. Mutations affecting the epigenetic machinery are expected to have widespread downstream epigenetic consequences, accounting for great pleiotropy of the genetic defect. The most frequent clinical manifestation is intellectual disability, suggesting that maintenance of the normal epigenotype is important for neuronal homeostasis. A wide variety of additional anomalies can occur, including limb malformations, disorders of the neuronal migration, immune dysfunction, growth impairment, and skeletal anomalies[8].
In this group, we find both well-known syndromes and little known syndromes. The latter include WDSTS, defined as such in 2010 by Koenig et al[11]. This syndrome has clinical features common to other chromatinopathies, but also quite peculiar features, which can lead to clinical suspicion, which then has to be confirmed by molecular tests. Because of the overlap of clinical features with other entities, a large panel of genes related to chromatinopathies should be taken into consideration. With the introduction of the most sophisticated diagnostic techniques of molecular genetics, such as next generation sequencing, the clinician can even have a diagnostic indication without suspecting a specific pathology[49]. This is a great opportunity, but it should not prompt the clinician to neglect the refinement of diagnostic skills, which can now also be supported by new technological aids[7].
At a time when there is a growing interest in epigenetics and in how genetic expression is controlled by chromatin remodeling, the study of chromatinopathies can provide useful elements to improve our knowledge, with very important repercussions both in the field of physiology, pathology, and aging[50].
| 1. | Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7446] [Cited by in RCA: 7810] [Article Influence: 411.1] [Reference Citation Analysis (0)] |
| 2. | Bártová E, Krejcí J, Harnicarová A, Galiová G, Kozubek S. Histone modifications and nuclear architecture: a review. J Histochem Cytochem. 2008;56:711-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 232] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 3. | Längst G, Manelyte L. Chromatin Remodelers: From Function to Dysfunction. Genes (Basel). 2015;6:299-324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 4. | Tyagi M, Imam N, Verma K, Patel AK. Chromatin remodelers: We are the drivers!! Nucleus. 2016;7:388-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 5. | Bjornsson HT. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473-1481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 6. | Fahrner JA, Bjornsson HT. Mendelian disorders of the epigenetic machinery: tipping the balance of chromatin states. Annu Rev Genomics Hum Genet. 2014;15:269-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 120] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 7. | Pascolini G, Fleischer N, Ferraris A, Majore S, Grammatico P. The facial dysmorphology analysis technology in intellectual disability syndromes related to defects in the histones modifiers. J Hum Genet. 2019;64:721-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 8. | Zollino M, Lattante S, Orteschi D, Frangella S, Doronzio PN, Contaldo I, Mercuri E, Marangi G. Syndromic Craniosynostosis Can Define New Candidate Genes for Suture Development or Result from the Non-specifc Effects of Pleiotropic Genes: Rasopathies and Chromatinopathies as Examples. Front Neurosci. 2017;11:587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 9. | Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589-3599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 480] [Cited by in RCA: 443] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 10. | Kim JH, Lee JH, Lee IS, Lee SB, Cho KS. Histone Lysine Methylation and Neurodevelopmental Disorders. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 11. | Koenig R, Meinecke P, Kuechler A, Schäfer D, Müller D. Wiedemann-Steiner syndrome: three further cases. Am J Med Genet A. 2010;152A:2372-2375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Wiedemann HR, Kunze J, Grosse FR, Dibbern H, Atlas of Clinical Syndromes: A Visual Aid to Diagnosis for Clinicians and Practicing Physicians. A syndrome of abnormal facies, short stature, and psychomotor retardation. Atlas of Clinical Syndromes: A Visual Aid to Diagnosis for Clinicians and Practicing Physicians. 2nd ed. London: Wolfe Publishing Ltd. (pub.) 1989; 198-199. |
| 13. | Steiner CE, Marques AP. Growth deficiency, mental retardation and unusual facies. Clin Dysmorphol. 2000;9:155-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Flannery DB, Fink SM, Francis G, Gilman PA. Hypertrichosis cubiti. Am J Med Genet. 1989;32:482-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 17] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Edwards MJ, Crawford AE, Jammu V, Wise G. Hypertrichosis "cubiti" with facial asymmetry. Am J Med Genet. 1994;53:56-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Visser R, Beemer FA, Veenhoven RH, De Nef JJ. Hypertrichosis cubiti: two new cases and a review of the literature. Genet Couns. 2002;13:397-403. [PubMed] |
| 17. | MacDermot KD, Patton MA, Williams MJ, Winter RM. Hypertrichosis cubiti (hairy elbows) and short stature: a recognisable association. J Med Genet. 1989;26:382-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 18. | Polizzi A, Pavone P, Ciancio E, La Rosa C, Sorge G, Ruggieri M. Hypertrichosis cubiti (hairy elbow syndrome): a clue to a malformation syndrome. J Pediatr Endocrinol Metab. 2005;18:1019-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Koç A, Karaer K, Ergün MA, Cinaz P, Perçin EF. A new case of hairy elbows syndrome (hypertrichosis cubiti). Genet Couns. 2007;18:325-330. [PubMed] |
| 20. | Jones WD, Dafou D, McEntagart M, Woollard WJ, Elmslie FV, Holder-Espinasse M, Irving M, Saggar AK, Smithson S, Trembath RC, Deshpande C, Simpson MA. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am J Hum Genet. 2012;91:358-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 227] [Article Influence: 16.2] [Reference Citation Analysis (1)] |
| 21. | Strom SP, Lozano R, Lee H, Dorrani N, Mann J, O'Lague PF, Mans N, Deignan JL, Vilain E, Nelson SF, Grody WW, Quintero-Rivera F. De Novo variants in the KMT2A (MLL) gene causing atypical Wiedemann-Steiner syndrome in two unrelated individuals identified by clinical exome sequencing. BMC Med Genet. 2014;15:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (1)] |
| 22. | Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 831] [Cited by in RCA: 834] [Article Influence: 34.8] [Reference Citation Analysis (3)] |
| 23. | Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 577] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 24. | Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, Barsyte-Lovejoy D, Al-awar R, Katona BW, Shilatifard A, Huang J, Hua X, Arrowsmith CH, Berger SL. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 394] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 25. | Kim SY, Levenson JM, Korsmeyer S, Sweatt JD, Schumacher A. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. J Biol Chem. 2007;282:9962-9972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Jakovcevski M, Ruan H, Shen EY, Dincer A, Javidfar B, Ma Q, Peter CJ, Cheung I, Mitchell AC, Jiang Y, Lin CL, Pothula V, Stewart AF, Ernst P, Yao WD, Akbarian S. Neuronal Kmt2a/Mll1 histone methyltransferase is essential for prefrontal synaptic plasticity and working memory. J Neurosci. 2015;35:5097-5108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 27. | Sun Y, Hu G, Liu H, Zhang X, Huang Z, Yan H, Wang L, Fan Y, Gu X, Yu Y. Further delineation of the phenotype of truncating KMT2A mutations: The extended Wiedemann-Steiner syndrome. Am J Med Genet A. 2017;173:510-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (1)] |
| 28. | Steel D, Salpietro V, Phadke R, Pitt M, Gentile G, Massoud A, Batten L, Bashamboo A, Mcelreavey K, Saggar A, Kinali M. Whole exome sequencing reveals a MLL de novo mutation associated with mild developmental delay and without 'hairy elbows': expanding the phenotype of Wiedemann-Steiner syndrome. J Genet. 2015;94:755-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 29. | Dunkerton S, Field M, Cho V, Bertram E, Whittle B, Groves A, Goel H. A de novo Mutation in KMT2A (MLL) in monozygotic twins with Wiedemann-Steiner syndrome. Am J Med Genet A. 2015;167A:2182-2187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 30. | Miyake N, Tsurusaki Y, Koshimizu E, Okamoto N, Kosho T, Brown NJ, Tan TY, Yap PJ, Suzumura H, Tanaka T, Nagai T, Nakashima M, Saitsu H, Niikawa N, Matsumoto N. Delineation of clinical features in Wiedemann-Steiner syndrome caused by KMT2A mutations. Clin Genet. 2016;89:115-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 31. | Mendelsohn BA, Pronold M, Long R, Smaoui N, Slavotinek AM. Advanced bone age in a girl with Wiedemann-Steiner syndrome and an exonic deletion in KMT2A (MLL). Am J Med Genet A. 2014;164A:2079-2083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Li N, Wang Y, Yang Y, Wang P, Huang H, Xiong S, Sun L, Cheng M, Song C, Cheng X, Ding Y, Chang G, Chen Y, Xu Y, Yu T, Yao RE, Shen Y, Wang X, Wang J. Description of the molecular and phenotypic spectrum of Wiedemann-Steiner syndrome in Chinese patients. Orphanet J Rare Dis. 2018;13:178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 33. | Bramswig NC, Lüdecke HJ, Alanay Y, Albrecht B, Barthelmie A, Boduroglu K, Braunholz D, Caliebe A, Chrzanowska KH, Czeschik JC, Endele S, Graf E, Guillén-Navarro E, Kiper PÖ, López-González V, Parenti I, Pozojevic J, Utine GE, Wieland T, Kaiser FJ, Wollnik B, Strom TM, Wieczorek D. Exome sequencing unravels unexpected differential diagnoses in individuals with the tentative diagnosis of Coffin-Siris and Nicolaides-Baraitser syndromes. Hum Genet. 2015;134:553-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 34. | Feldman HR, Dlouhy SR, Lah MD, Payne KK, Weaver DD. The progression of Wiedemann-Steiner syndrome in adulthood and two novel variants in the KMT2A gene. Am J Med Genet A. 2019;179:300-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 35. | Min Ko J, Cho JS, Yoo Y, Seo J, Choi M, Chae JH, Lee HR, Cho TJ. Wiedemann-Steiner Syndrome With 2 Novel KMT2A Mutations. J Child Neurol. 2017;32:237-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | Stellacci E, Onesimo R, Bruselles A, Pizzi S, Battaglia D, Leoni C, Zampino G, Tartaglia M. Congenital immunodeficiency in an individual with Wiedemann-Steiner syndrome due to a novel missense mutation in KMT2A. Am J Med Genet A. 2016;170:2389-2393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 37. | Enokizono T, Ohto T, Tanaka R, Tanaka M, Suzuki H, Sakai A, Imagawa K, Fukushima H, Iwabuti A, Fukushima T, Sumazaki R, Uehara T, Takenouchi T, Kosaki K. Preaxial polydactyly in an individual with Wiedemann-Steiner syndrome caused by a novel nonsense mutation in KMT2A. Am J Med Genet A. 2017;173:2821-2825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 38. | Baer S, Afenjar A, Smol T, Piton A, Gérard B, Alembik Y, Bienvenu T, Boursier G, Boute O, Colson C, Cordier MP, Cormier-Daire V, Delobel B, Doco-Fenzy M, Duban-Bedu B, Fradin M, Geneviève D, Goldenberg A, Grelet M, Haye D, Heron D, Isidor B, Keren B, Lacombe D, Lèbre AS, Lesca G, Masurel A, Mathieu-Dramard M, Nava C, Pasquier L, Petit A, Philip N, Piard J, Rondeau S, Saugier-Veber P, Sukno S, Thevenon J, Van-Gils J, Vincent-Delorme C, Willems M, Schaefer E, Morin G. Wiedemann-Steiner syndrome as a major cause of syndromic intellectual disability: A study of 33 French cases. Clin Genet. 2018;94:141-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 39. | Aggarwal A, Rodriguez-Buritica DF, Northrup H. Wiedemann-Steiner syndrome: Novel pathogenic variant and review of literature. Eur J Med Genet. 2017;60:285-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 40. | Chen M, Liu R, Wu C, Li X, Wang Y. A novel de novo mutation (p.Pro1310Glnfs*46) in KMT2A caused Wiedemann-Steiner Syndrome in a Chinese boy with postnatal growth retardation: a case report. Mol Biol Rep. 2019;46:5555-5559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 41. | Stoyle G, Banka S, Langley C, Jones EA, Banerjee I. Growth hormone deficiency as a cause for short stature in Wiedemann-Steiner Syndrome. Endocrinol Diabetes Metab Case Rep. 2018;2018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 42. | Chan AJS, Cytrynbaum C, Hoang N, Ambrozewicz PM, Weksberg R, Drmic I, Ritzema A, Schachar R, Walker S, Uddin M, Zarrei M, Yuen RKC, Scherer SW. Expanding the neurodevelopmental phenotypes of individuals with de novo KMT2A variants. NPJ Genom Med. 2019;4:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 43. | Lebrun N, Giurgea I, Goldenberg A, Dieux A, Afenjar A, Ghoumid J, Diebold B, Mietton L, Briand-Suleau A, Billuart P, Bienvenu T. Molecular and cellular issues of KMT2A variants involved in Wiedemann-Steiner syndrome. Eur J Hum Genet. 2018;26:107-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 44. | Bogaert DJ, Dullaers M, Kuehn HS, Leroy BP, Niemela JE, De Wilde H, De Schryver S, De Bruyne M, Coppieters F, Lambrecht BN, De Baets F, Rosenzweig SD, De Baere E, Haerynck F. Early-onset primary antibody deficiency resembling common variable immunodeficiency challenges the diagnosis of Wiedeman-Steiner and Roifman syndromes. Sci Rep. 2017;7:3702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 45. | Zhang H, Xiang B, Chen H, Chen X, Cai T. A novel deletion mutation in KMT2A identified in a child with ID/DD and blood eosinophilia. BMC Med Genet. 2019;20:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 46. | Grangeia A, Leão M, Moura CP. Wiedemann-Steiner syndrome in two patients from Portugal. Am J Med Genet A. 2020;182:25-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 47. | Suleiman J, Riedhammer KM, Jicinsky T, Mundt M, Werner L, Gusic M, Burgemeister AL, Alsaif HS, Abdulrahim M, Moghrabi NN, Nicolas-Jilwan M, AlSayed M, Bi W, Sampath S, Alkuraya FS, El-Hattab AW. Homozygous loss-of-function variants of TASP1, a gene encoding an activator of the histone methyltransferases KMT2A and KMT2D, cause a syndrome of developmental delay, happy demeanor, distinctive facial features, and congenital anomalies. Hum Mutat. 2019;40:1985-1992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 48. | Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 212] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 49. | Lonardo F, Lonardo MS, Acquaviva F, Della Monica M, Scarano F, Scarano G. Say-Barber-Biesecker-Young-Simpson syndrome and Genitopatellar syndrome: Lumping or splitting? Clin Genet. 2019;95:253-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 50. | Zhang W, Qu J, Liu GH, Belmonte JCI. The ageing epigenome and its rejuvenation. Nat Rev Mol Cell Biol. 2020;21:137-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 324] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: European Society of Medical Genetics (Fellow); Società italiana di Genetica Umana (Italian Society of Human Genetics) (Fellow); and European Cytogeneticists Association (Fellow).
Specialty type: Genetics and heredity
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: de la Serna I, Hu JF, Zhe MM S-Editor: Tang JZ L-Editor: Wang TQ E-Editor: Liu JH