INTRODUCTION
Genome multiplication in the cells of placental trophoblast is a unique phenomenon among the mammalian and other vertebrate tissues. By now, ontogenesis of rodent trophoblast cells is fairly characterized and serves as a model for studying normal and pathological development of placenta. The main peculiarities of the trophoblast cell lifespan in the rodent placenta is a genome reproduction due to cell cycle reduction up to two phases-S and G, the multifold repeat of which results in high level of cell polyploidization-up to 512-2048 and higher[1-4] that involves polytenization[5]. Beginning from the second half of pregnancy, a significant part of the secondary giant trophoblast cells undergo depolytenization and genome segregation with subsequent isolation of numerous small nuclear fragments detaching from the giant nucleus[5-8].
A range of trophoblast cell populations do not leave mitotic cycle (a part of them probably represent the trophoblast stem cells); the cells and their derivatives form cell population of lower ploidy levels (2c-32c) at the account of uncompleted mitoses[5-7,9,10]. By now, a noticeable data are available in the polyploidy in the other mammalian groups, their modes of polyploidization being different from the rodent ones[11,12]. Simultaneously, the more and more data appear on the role of polyploidy in formation of different mammalian tissues[13-19]. In particular, in recent publications a great attention is drawn to the relationship of polyploidy and aneuploidy, the latter is considered as a factor of genetic variability that may be an adaptive under the stress conditions[17-21]. Therefore it seems to be interesting to compare regularities of genome multiplication in different mammalian species in accordance with possible role of trophoblast cells in placenta formation.
THE WAYS OF SOMATIC CELL POLYPLOIDIZATION
At present a great number of data has been obtained that confirm the concept that modification (mostly, shortening) of the “archetypal” mitotic cell cycle results in genome multiplication[3,7,14,19,22-24]. Recently, there dominates a notion that such a modified cell cycle is characterized by alternating DNA synthesis (S) and Gap (G) phases in the absence of intervening mitoses, karyokinesis, and cytokinesis; a series of these shortened cycles allows cells to achieve high level of ploidy that may exceed 1000c[19,23,24]. Nevertheless, the extent of cell cycle shortening that results in polyploidization appears to differ significantly in different cell types and taxa. Therefore, it seems to be important to present short characteristics of different ways of polyploidization of somatic cells of different animals, plants, and human.
Switching off the last step of mitosis-cytokinesis-may be the first step to polyploidy. In this case, binucleate cell is formed (2c × 2). In the next cell cycle, both nuclei, as a rule, enter mitosis synchronously due to what the uniform metaphase plate is formed. If the mitosis comes to the end, it results in two mononucleated cells with tetraploid nuclei. Alternation of acytokinetic and following complete mitoses may result in formation of mono- and binucleate cells of the higher ploidy:
2c-(2c × 2)-4c-(4c × 2)-8c-(8c × 2)
Noteworthy, in this case, mitosis is the key event that allows formation of polyploid nucleus. Such a way of polyploidization was demonstrated basing on the dynamics of transition of mono- and binucleate cells with nuclei of different ploidy by using combination of cytophotometry and 3H-thymidine DNA replication labeling[22]; this way was currently confirmed by using time-lapse video images[18]. Similar way of polyploidization was also demonstrated in some other mammalian cell types-cardiomyocytes[25,26] and aortic vascular smooth muscle cells[27].
Block of mitoses at the meta- and anaphase also may result in polyploidization: the cell do not complete the mitotic division owing to what the nucleus with the doubled number of chromosomes is formed[3,7]. Such a way of polyploidization recently is often described under the name of endomitosis[14,19,23]. However, taking into account participation of mitosis (although uncompleted) in this cell cycle, we adhere to the used term “restitutional” or “uncompleted mitosis”[3,28]. And now, in our opinion, it makes sence to unify the cases of uncompleted and acytokinetic mitoses under the term “reduced mitoses”. Meantime, the term “endomitosis” includes prefix “endo” accounted for by the initial sence that implied chromosome/chromatid segregation inside the nucleus. Different forms of reduced mitoses may be observed simultaneously in the same cell type; in some cases they are accompanied by endocycles (see below); all these phenomena may result from disorder of many mitotic events. A good case in point is aortic vascular smooth cells studied by the time-lapse video[27]. In most cases there occurred normal anaphase chromosome segregation, but progress of cytokinesis was arrested; as a result, binucleate cells were formed. Sometimes such a binucleate cell renewed cytokinesis that came to the end with two daughter (probably diploid) cells formation. In other, rare, cases, mitoses resulted in mononuclear polyploid cells due to chromosome bridge(s) that did not allow forming two daughter nuclei. At last, there occurred some mitoses with shallow cleavage furrow and missegregation of sister chromatids; thereafter cleavage furrow disappeared and mononuclear polyploid cell was formed. All the phenomena were accompanied by downregulation of Survivin[27]; the disturbance of AuroraB/Survivin complex (a regulator of mitotic machinery) exerted such a pleiotropic effect on the progression of mitosis. The pleiotropic modifications of mitosis including endocycles (see below) were also observed in many cases of spontaneous and induced polyploidization, in particular, in endosperm and suspensor of higher plants[3,28], in cancer cells[29], in bovine trophoblast cells[11,30] and in some other cases.
The reduced mitoses in most cases can result in polyploidy of moderate level such as 4c, 8c, and some higher. As a rule, these modes of polyploidization allow cell to retain its mitotic potential[17,18]. In some cases they may result in “ploidy reversal” thereby generating diploid cells from the tetra- and octaploid ones as demonstrated by the time-lapse video[17,18,20].
More profound reduction of mitotic cycle results in endocycles. Originally this term covered all cases of genome multiplication accomplished without the nuclear envelope disappearance[3,5-7,28,31-33]. Therefore, it may be suggested that in these cases, everything or nearly all stages of mitosis are reduced: the chromosomes do not form the metaphase plate, and nucleus retains the traits of interphase or prophase. The giant cells with polytene chromosomes of salivary glands of Diptera that probably represent numerous copies of the tightly attached elongated sister chromatids with clear-cut chromomere structure most probably are formed by means of block of mitosis in prophase.
The cycle of polytene nucleus devoid of mitosis was described in the giant trophoblast cells of mouse and rat. Using 3H-thymidine labeling, two phases-S (endointerphase) and G (endoprophase) were identified in rat trophoblast cells. At the endointerphase (S-phase) the nucleus was filled with thin long paired Feulgen-positive threads, whereas at the endoprophase the bundles of non-classic polytene chromosomes were observed[5-7]. In mice, three phases of endoreduplication-G1, S, and G2 were discerned basing on the oscillative expression of the S-phase inhibitor p57kip2[34].
The so-called classic endomitosis costs independently from the point of view of classifications used here. It is also a cell cycle devoid of phase of mitosis, although it includes phases of endoprophase, endometaphase, and endoanaphase, when the chromosomes attached to the nuclear envelope undergo condensation and splitting into sister chromatids followed by decondensation[3,7,31,35]. Interestingly, the nuclear envelope in most cases is retained throughout the whole cycle of endomitosis. Using the 3H-thymidine delayed labeling, three phases of endointerphase-G1, S, and G2 were determined in the endomitotic cells of albumen gland of the snail Succinea lauta[36]. All the data, in our opinion, suggest that classic endomitosis, most likely, belongs to endocycles, i.e., modifications of cell cycle, in which the genome multiplication is accomplished due to reduction of most of mitotic stages.
The classic and some other types of endomitosis are widespread mostly in invertebrates and plant tissues[3,28]. Meanwhile, such a mode of genome multiplication is observed in the human placental trophoblast[7,32,37,38], as well as in cancer cells[29,33,37]. The future investigation should elucidate the preferable meaning of the term “endomitosis”.
Numerous data suggest that endocycles allow cells to gain very high ploidy levels. Thus, a series of endoreduplication cycle polytene nuclei of Drosophila salivary glands may result in 1024c[3,39-41] that most frequently provides irreversibility of switch from mitotic cycle to endoreduplication[22,23]. That is why, probably, endocycles are characteristic of the majority of invertebrate and plant differentiated tissues[3,28,42]. By contrast, the uncompleted mitoses including the acytokinetic ones in most cases can result in polyploidy of moderate level such as 4c, 8c, and some higher. As a rule, these modes of polyploidization allow cells to retain their mitotic potential-for example, in hepatocytes[17,18] that enable tissue to undergo an effective regeneration and sometimes (mostly in cases of experiments), to reverse to the diploid state[17,18].
POLYPLOIDIZATION IN DIFFERENTIATION OF THE RAT AND MOUSE TROPHOBLAST CELLS
Beginning from the onset of differentiation, the primary and secondary trophoblast giant cells (TGC) in rat and mouse placenta undergo a series of endoreduplication cycles that result in a very high ploidy level[1,4,6,43] and polytenization of giant nuclei. In the non-classic polytene nuclei, chromatin underwent condensation in the G-phase showing bundles of thick and short chromonemes under the nuclear envelope or near the nucleolus and decondensation in the S-phase where nuclei were similar to the interphase ones[5,6]. Later on, i.e., after the 12th gestational day in rat, the features of non-classic polytenic chromosomes become less expressed that the precede transformation of the giant nucleus into polygenomic and multinucleate[5]. Switch to endoreduplication in the murine TGC is connected to the switch of the cyclin D isoform expression from D3 to D1; the arrest of mitotic cycle and the onset of endoreduplication was most probably accounted for by the failure to assemble the cyclin B/p34cdk1 complex during the first endocycle; in the subsequent endocycle the mitotic cyclin B was suppressed[43,44].
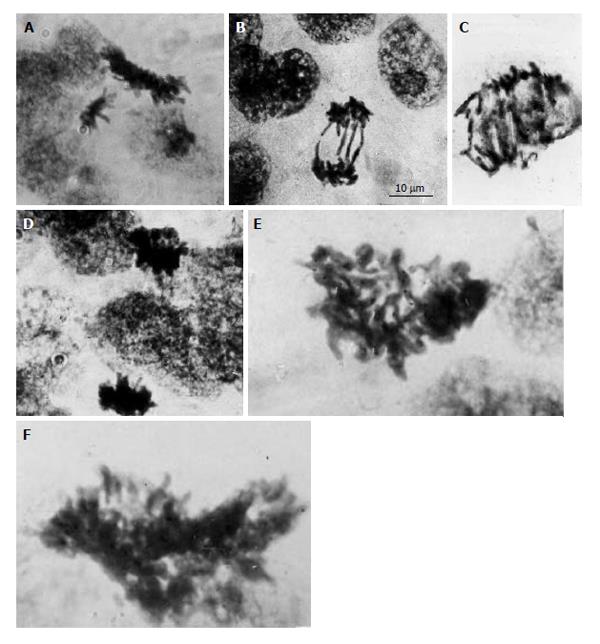
As distinct from the primary and secondary TGC, the junctional zone (JZ) trophoblast cells in rat and mouse placenta represent a highly proliferative cell population; the mitotic activity of JZ trophoblast cells persists up to midgestation[6]. Simultaneously the JZ trophoblast cells undergo differentiation into a number of cell subtypes involved in glycogen store, hormone production, invasion, etc.[9,45-49]. Unlike the secondary TGC that form a barrier at the border between semiallogenic maternal and embryonic tissues, the JZ trophoblast cells undergo polyploidization via uncompleted polyploidizing mitoses up to 8c followed by some endoreduplication cycles allowing them to reach 16-32c[6]. The DNA content measurement in the mitotic figures showed 4c, 8c and 16c mitotic figures indicating the ability of 2c-8c polyploid cells to undergo the complete mitotic division[6]. However, the polyploid mitotic figures often were of abnormal shape; chromosome bridges were frequent in polyploid anaphases, the double and multiple bridges were commonly present (Figure 1). Therefore, in this case, polyploidization increases the possibility of mitotic arrest that may result in interphase renewal leading to further polyploidization.
Figure 1 Diploid and polyploid mitoses in the junctional zone trophoblast cells in rat placenta at the 14th day of gestation.
A: Tetraploid metaphase and diploid anaphase; B: Diploid restitutional anaphase with multiple chromosome bridges; C: Polyploid restitutional anaphase with multiple chromosome bridges; D: Tetraploid normal anaphase; E: Octaploid metaphase; F: Hexadecaploid metaphase.
Difference in endocycle and uncompleted mitosis is illustrated by the data on their regulation by the transcription factor family E2F[24,50]. The canonical and atypical transcriptional programs converge to control the endocycle through the regulation of cellular events important for mitosis, karyokinesis, and cytokinesis. Thus, in the murine trophoblast giant cells (TGC) the targeted gene inactivation of E2f1, E2f2, and E2f3 transcription induced abnormally large nuclei, their ploidy exceeding their characteristic ploidy levels 8c-256c. In the E2F7-/- and E2F8-/- an unusual number of mitosis at different phases were observed, which suggest interruption of endocycle[24]. The ploidy did not exceed 64c that, according to our data is normally characteristic of the trophoblast cells of rat placenta junctional zone undergone polyploidization via polyploidizing mitoses with subsequent endocycles[6]. This phenotype of E2F7-/- and E2F8-/- mice included upregulation of mitotic cyclins A2 and B1 in TGC as well as mitotic marker P-H3. In addition, up to 40% of nuclei underwent separation of two approximately equal nuclei out of the single giant nucleus[24]. The data suggest that the E2F7/E2F8 ablation promotes switch on mitotic cycle that restricts endocycling in TGC.
In liver, the combined ablation of E2f1, E2f2, and E2f3 resulted in an increase of ploidy level[24] that suggests a possibility of hepatocyte ectopic endocycling. By contrast, ablation of E2F7/E2F8 leads to hepatocyte diploidization and to a decrease of the binuclear cell number. Thus, the canonical and atypical E2Fs exerts opposite effect: the canonical ones promote complete progression of mitosis, whereas the ancient, atypical one increases endocycling. Interestingly, study of the global gene expression in hepatocytes showed that majority of genes upregulated in E2F7/E2F8-deficient hepatocytes were downregulated in the E2F1/E2F2/E2F3-deficient ones, many of these genes had annotated to be the cell cycle function related, in particular, to bound to the G2/M transition or to mitosis[24].
Loss of E2F1 suppressed some, but not all mitotic defects in TGC and hepatocytes caused by E2F7/E2F8-deficiency. Ablation of mitotic cyclins in the E2F7/E2F8-deficient mice resulted in the greater ploidy in TGC, whereas hepatocytes in mice of similar phenotype showed similar ploidy as wild type mice. Thus, inhibiting the transcriptional network that signals G2/M progression or interfering with mitotic machinery (by cyclins A1/A2 ablation) reinstablished the mitotic block and reinitiated higher ploidy levels[24], probably, via endocycle progression. The data demonstrate that an intricate E2F network involving balanced and antagonistic activities of canonical (E2F1-3) activators and atypical (E2F7/E2F8) repressors plays, most probably, one of the key roles in the mammalian endocycle control. Perhaps, most dramatic manifestation of altering the balance in the E2F network is ectopic mitoses in the E2F7/E2F8 deficient TGC. Interestingly, such a phenotype was lethal, whereas inactivation of these genes did not produce any effect on the growth of liver. Thus, in the liver, mitosis is a prerequisite of polyploidization whereas in the highly endopolyploid murine TGC, in distinct from liver, such a key event is almost complete reduction of mitosis (i.e., endoreduplication). It may be important for some functional peculiarities of these cell types: for example, retention of mitotic potential may be important for maintenance of regenerative ability of the liver.
Difference in the cell cycle machinery was also detected by using transcriptome analysis of the murine TGC and megakaryocytes[51]. The authors compared the expression level of orthologous genes in the DNA replication pathway and cell cycle gene controlling the G1-S transition and S-phase. The components of the DNA replication machinery including the origin recognition complex, minichromosome maintenance and proliferating cell nuclear antigen genes were strongly expressed in the TGC lineage. In contrast, polyploid megakaryocytes exhibited a reduction in the expression of these genes. The TGC similar to Drosophila polytene cells, on the contrary, showed a reduction in the expression of mitotic genes, as compared with diploid control embryonic cells[51], thereby confirming endocycle origin of their high level of ploidy[5,34,42]. Meantime, megakaryocytes showed increased expression of M-phase factors as compared to TGC or Drosophila[51]. Such a comparison suggests that genes controlling G1-S transition might play a key role in genome multiplication via endoreplication (in murine trophoblast and Drosophila salivary glands). By contrast, upregulation of mitotic events rather than S-phase factors play a key role in genome multiplication of megakaryocytes via endomitosis (reduced mitosis).
Significance of the two different ways of genome multiplication-endoreplication and reduced mitoses-can be shown on an example of functional organization of different trophoblast cell populations in the rodent placenta. According to our recent data, different trophoblast cell population show different patterns of intracellular cytokeratin localization. Long massive cytokeratin threads were found in the peripheral cytoplasm of the secondary TGC of rat and in their long sprouts by which they connected each other making continuous barrier at the border with decidua[7]. Similar cytokeratin immunostaining was observed in the trabecular spongiotrophoblast cells that line maternal blood sinuses. Clusters of low-ploidy proliferative and/or glycogen cells in the depth of JZ showed some weaker cytokeratin signal. Thus, the specific structure of giant trophoblast cells seems to provide a barrier between semiallogenic fetal and maternal tissues.
The east-european field vole Microtus rossiaemeridionalis provides another example of barrier function of TGC[52]. At the beginning of placentation TGC also form a continuous layer at the foeto-maternal interface. However, at midgestation, clusters of tightly attached low-ploid glycogen-rich junctional zone trophoblast cells progressively replace TGC thereby drawing them in the depth of the fetal part of placenta. Nevertheless, TGCs bound by their heavily cytokeratin-positive sprouts form a framework that holds other trophoblast cell populations and line lacunae with maternal blood.
Significance of specific TGC organization is confirmed by the data on the compound mutants on Cytokeratin 8 and 19[53]. In this case, there was an excessive number of TGC that however were not tightly attached to each other. Besides, K8-/-, K8-/-/K19-/- and K18-/-/K19-/- knockout conceptuses died by the moment of placenta formation and showed placental hemorrhaging[54-56]. This apparently caused flooding directly to fetal tissue where these trophoblast cells normally separate embryonic blood from maternal circulation.
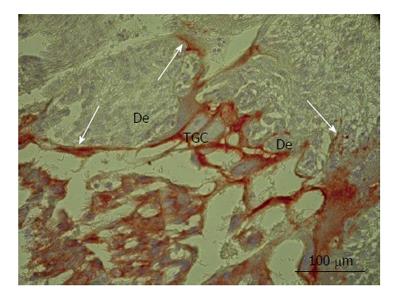
The enormous sprouts of TGC probably play another role in rat placenta. Some secondary TGC protrude the decidual tissue by means of large nippel-like highly cytokeratin-positive sprouts (Figure 2) that surround wide accumulations of decidual cells, the latter, probably, undergoing subsequent degradation[54]. This process was observed throughout the most of pregnancy and represents, most probably, a special kind of invasion that involves phagocytosis of decidual tissue by the trophoblast that may be called “group phagocytosis”. The trophoblast phagocytosis that provides histotrophic nutrition of embryo was described in detail in the endothelio- and hemochorial placenta of a range of mammalian species[6,57-59]. Thus, in mice, processes of trophoblast giant cells penetrate a layer of uterine epithelial cells and internalize the cells[58]; phagosomes with fragments of decidual cells are observed in TGC. Recently, transcriptome analysis showed high expression of scavenger receptor class B, member 1 (scarb 1) required for phagocytic activity of TGC[51]. Therefore, gigantism of TGC in this case probably allow them to perform invasion as well as partial degradation and phagocytosis of decidualized endometrium. It is interesting to note that one of the possible advantages of endoreduplication is that cell growth is accomplished without cell division that would imply a significant rearrangement of cytoskeleton[23]. Mitotic proliferation would prevent establishment of such a continuous system of the tightly attached phagocytosing TGC. Therefore, the endoreduplication allows TGC to combine growth at the restricted time period with formation of a barrier made of the tightly attached TGC that protect embryo from the immunological attack of the maternal organism and provide its histotrophic nutrition. It is also notable that the endoreduplication allows TGC growing without nuclear envelope disappearance. It also may be important, because isolation of genome inside the nuclear envelope may protect it from mutagenic effect of the degrading DNA of the phagocytosed maternal cells. In rare cases, in the field vole placenta we observed erythrocyte, i.e., the anuclear cell, inside the phagocytic trophoblast cell undergone mitosis[6].
Figure 2 Secondary trophoblast giant cells of rat placenta at the 16th day of gestation.
Note massive long cytokeratin-positive sprouts that embrace wide zones of decidual tissue (De). TGC: Trophoblast giant cells.
An interesting example of the necessity of endopolyploidy for the “barrier” function is the recently obtained data on subperineural glia (SPG) of Drosophila melanogaster by Italic. These glial cells were highly polyploid, and ploidy correlated with brain mass. Inhibition of the SPG polyploidy caused rupture of the septate junctions necessary for the blood-brain barrier. Thus, the increased SPG cell size resulting from polyploidization is required to maintain the SPG envelope surrounding the growing brain[60].
As to the trophoblast cell populations lying in the depth of the fetal part of placenta that first represent a proliferative pool of trophoblast, their primary steps of genome multiplication through reduced mitoses probably allow them to accumulate the great number of cells undergoing multidirectional differentiation into a number of subsets of cells within JZ and subsequently migrating into decidua (endovascularly and interstitially) as well as consisting trophoblast of labyrinth[6,7,61-63].
GENOME SEGREGATION: A TERMINAL STEP OF RODENT TROPHOBLAST GIANT CELL LIFESPAN
Beginning from the second half of pregnancy, the secondary giant trophoblast cells in rodent placenta undergo the so-called nuclear fragmentation when a part of giant nuclei break down into a number of the smaller nuclear fragments[5,7,64-66]. The fragmentation is preceded by depolytenization which is gained by disintegration of polytene chromosome bundles into the double chromonemes/endochromosomes; the process was described in detail earlier[5]. A similar process was recently investigated by using in situ hybridization the whole-chromosome labeling in the polytene chromosomes of Calliphora erythrocephala ovarian nurse cells[67]. In rat placenta, a portion of TGC undergoing nuclear fragmentation increases as the end of pregnancy approaches. Noteworthy, attenuation of DNA replication precedes nuclear fragmentation: this process begins in the nuclei that lost their capability for DNA synthesis[64]. Nevertheless, there were some reports about scarce murine 3H-thymidine-labeled giant trophoblast cell nuclei undergoing fragmentation[66].
Interestingly, the process of nuclear fragmentation often begins with break down of the giant nuclei into two approximately equal parts (Figure 3). As a rule, one of these “parts” (“subnuclei”) undergoes more complete breakdown into numerous fragments. It is notable that ablation of mitosis progression regulators E2F1, E2F2, and E2F3 often resulted in break-down of mouse trophoblast giant nuclei into two approximately equal parts[24]. This suggests that although the process of giant nuclear segregation is not similar to mitosis, most probably, some elements of the mitotic machinery are involved in this mechanism.
Figure 3 Fragmentation of the rat trophoblast giant cells.
A and B: First the giant nuclei fall into two large nuclei, the latter then breaks down into numerous small fragments (Fr); B: The giant nucleolus falls down into several nucleoli (Nl), then they form small nucleoli that seem to move into small nuclear fragments, some nucleoli in the nuclear fragments may be formed de novo. Heidenhain hematoxylin staining.
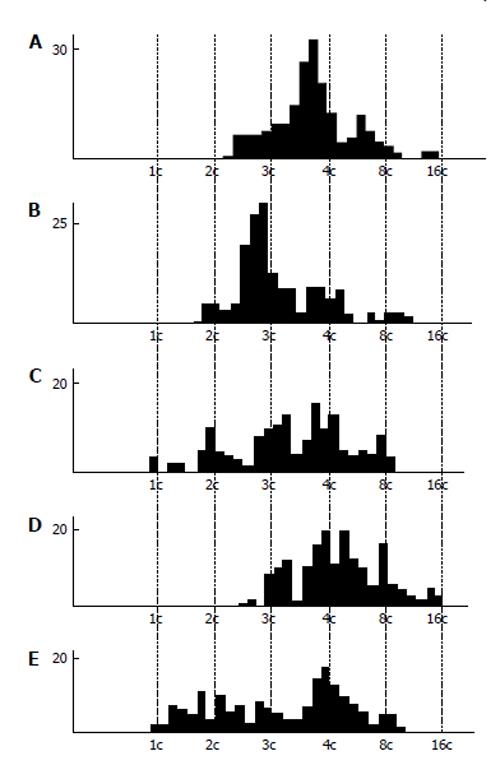
Cytophotometric and cytofluorometric measurement of DNA content measurement showed a tendency to the whole-genome distribution of DNA into the nuclear fragments (Figure 4). In the rat secondary TGC, the histograms show quite clearly distinguishable peaks corresponding to different ploidy classes, i.e., 1c, 2c, 4c, 8c, 16c, and 32c. However, there occurred incidence of some intermediate values of DNA content that could not be explained by the DNA synthesis, because in the nuclear fragments the S-phase is absent. In the mouse secondary TGC the tendency for the whole-genome distribution was also observed, but some peaks tend to be 3c and 6c values[5,64]. Similar data were obtained on the giant cells of the rabbit trophoblast[5].
Figure 4 DNA content in the nuclear fragments of secondary trophoblast giant cells of mouse (A and B) and rat (C-E) at the 17th (A), 19th (B), 14th (C), 16th (D), and 18th (E) day of gestation.
Abscissa: The DNA content (arbitrary units, logarithmic scale), and ploidy, c; Ordinate: the number of nuclear fragments.
The DNA content measurement of nuclear fragments of the field vole secondary giant trophoblast cells showed more clear-cut correspondence to the distinct ploidy classes multiple to 2c[5,68].
Behavior of natural chromosome markers, i.e., sex chromatin body and nucleolus, were observed in the giant nuclei undergoing fragmentation. The inactivated X-chromosome that forms a condensed chromatin body in the interphase nucleus was undergone to successive doubling in each cycle on endoreduplication of the rat secondary TGC of female embryos[1]. In the course of fragmentation, each nuclear fragment obtained a small condensed sex chromatin body attached to the nuclear envelope[5,65]. Similarly, each fragment contains a small nucleolus. At the onset of fragmentation, when the nucleus breaks down into several rather large fragments, the large nucleoli also break down into several ones that seem to be converted into separate fragments (Figure 3). Nevertheless, as to the final steps of fragmentation, it is not easy to decide whether the small nucleoli came from the initial giant nucleus or were synthesized de novo.
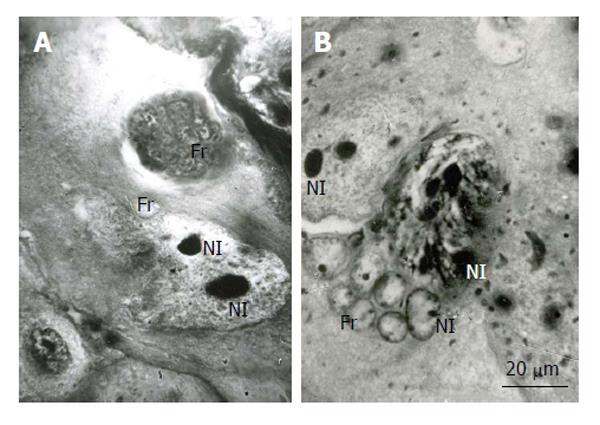
In the mouse secondary TGC, polytene chromosomes were characterized by the presence of large clear-cut heterochromatin blocks attached to the nucleolus (Figure 5). In the course of nuclear fragmentation, the heterochromatin blocks were separated into small blocks that were observed in the nuclear fragments near the nucleolus (Figure 5).
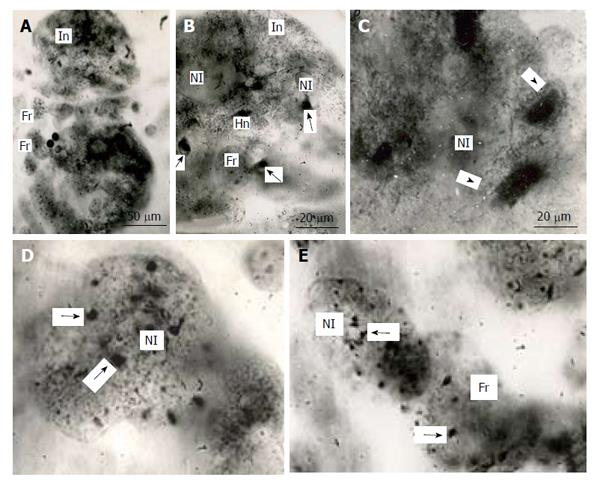
Figure 5 Distribution of heterochromatin blocks into nuclear fragments of mouse trophoblast giant cells.
A: The initial nucleus (In) of trophoblast giant cells (TGC) in the process of fragmentation; B: The initial nucleus contains large clear-cut heterochromatin blocks (Hn, arrows) near nucleoli (Nl); C: The nucleus contains non-classic polytene chromosomes with numerous distal loops (arrowheads); D and E: Nuclear fragments with small heterochromatin blocks (arrows) near nucleoli. Squash preparations, aceto-orcein staining. Fr: Nuclear fragments.
To determine whether the separate chromosomes are distributed into the nuclear fragments in correspondence with ploidy level, we used the so-called gonosomal chromatin bodies (GCB) in the secondary TGC of the field vole Microtus rossiaemeridionalis by Italic that represent large heterochromatin blocks of X and Y chromosomes that form prominent condensed chromatin bodies in the interphase nucleus. According to the DNA content measurement in the nuclear fragments of the secondary TGC and their GCBs, each fragment contains GCB(s) in correspondence with its ploidy level[8].
The above-mentioned data confirm that isolation of small “nuclei” inside the TGC is a process in which single chromosomes are regularly dispersed into nuclear fragments. Nevertheless, the data of cytophotometry leave open the possibility of some aneuploidy in the nuclear fragments. Thus, we can conclude that polytene nuclei of TGC are transformed into the multinucleate giant cells, each of nuclei contains the euploid (1c, 2c, 3c, 4c, etc.) or the near-euploid set of endochromosomes.
The nuclear fragmentation seems to be a rare common process, and its mechanism is not elucidated in detail. Active participation of membranes of the nuclear envelope and its derivatives was observed in the rat secondary TGC[64]; bundles of intermediate filaments were also observed in invagination of nuclear envelope of nuclei into the process of fragmentation. However, such a process, most probably, makes possible separation of one or more genomes into separate fragments, and their number is not always divisible (2n)c.
In the literature, very little data are known to draw analogies that could make an explanation of the above-mentioned chromosome distribution in nuclear fragments. Thus, in endomitotic nuclei of the seminal follicle wall of locust separation of some endochromosome groups corresponding to haploid, triploid, diploid, and hexadecaploid chromosome sets[69], i.e., segregation of a number of genomes not multiple to (2n)c. The data also confirm the possibility of the spatial separation of genomes without their entering mitosis.
Another example of genome segregation is observed in polyploid nuclei of protists. In some Radiolarians, nuclei are polyploid, and nuclear division is achieved via genome segregation[70]. For example, in Aulacantha by Italic, there is a single polyploid, the so-called primary nucleus. In the course of nuclear division, a great number of large chromosomes are presumably gathered into chains. During endomitosis that precedes division, reproduction of the “gathered” chromosomes takes place. In the course of sporogenesis, fragmentation of the primary nucleus results in a number of secondary nuclei; the latter initially lie in the same cytoplasm, but later on the cytoplasm also undergoes subdivision into a number of secondary bolls. Then the secondary nuclei undergo a series of division and, as a result, the cytoplasm breaks down into mononuclear prespores that divide once again to give rise to zoospores. All this complicated process is considered as a breakdown of the primary nucleus into separate genomes (via their segregation) and depolyploidization is carried out in several steps.
Since placenta is a provisory organ, it is felt that there should exist a mechanism of death of its cells, especially the peripheral ones) as the term approaches; it would enable placenta to separate at birth. In fact, the lifespan of the secondary rat TGC is strictly 22 d, which coincides with the length of pregnancy; it is not changed in culture and under conditions of transplantation under the kidney capsule[4].
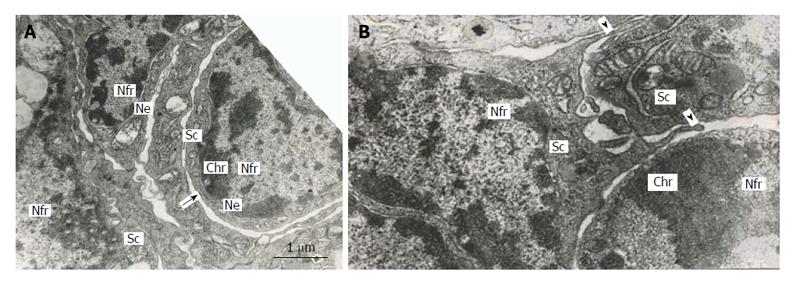
The multinucleate secondary TGC in rat placenta show some signs of apoptosis, attenuation of cell functions and degradation: condensation of chromatin located under the nuclear envelope of the nuclear fragments, inactivation of nucleolus, destruction of cytoplasmic organelles, etc.[5,7]. Nevertheless, the outer nuclear membrane of some nuclear fragments generates outgrowths continuous with the agranular endoplasmic reticulum, which, in turn, produces the double-membrane channels that delimit cytoplasmic mic territories around nuclear fragments. It is notable that the territories contain the whole set of cell organelles-mitochondria, Golgi complex, granular and agranular endoplasmic reticulum, numerous polysomes. In many cases, nuclear fragments with their cytoplasmic territories that can be called “subcellular compartments”, look quite viable; they are often isolated from the rest of cytoplasm of TGC that show signs of degradation. Moreover, sometimes this process results in isolation of subcellular compartments from other ones; however, in some cases the compartments connect each other by means of typical intercellular junctions (Figure 6). Occasionally it is possible to see as the pseudopodia formed by one subcellular compartment as though try to surround another one (Figure 6). Therefore, the subcellular compartments of TGC may behave like full value cells[7]. Meantime, in other cases, these compartments look apoptotic.
Figure 6 Subnuclear compartments in the rat trophoblast giant cells are separated from each other by forming rather wide channels of endoplasmic reticulum (A, arrow); some of them show intercellular junctions (A and B), some compartments (B) produce pseudopodia-like outgrowths (arrowheads) moving to other compartments.
Nfr: Nuclear fragments; Chr: Condensed chromatin; Sc: Subnuclear compartments; Ne: Nuclear envelope.
Thus, as the term approaches, TGC undergoing attenuation of reproductive, transcriptional, and other function simultaneously separates numerous subcellular compartments with near-euploid nuclei; the compartments may be both viable and apoptotic. It seems to be important that these fragments lose completely their capability for genome reproduction that would prevent renewal of proliferation of trophoblast cells. Nevertheless, the subcellular compartment formation may represent a reserve mechanism that preserve trophoblast genome for unknown functions. Further investigation may probably shed light on the significance of this phenomenon.
POLYPLOIDIZATION OF SILVER FOX
In the silver fox the trophoblast invasiveness is manifested in other form as compared to rodents. In Carnivores, syncytiotrophoblast only partially destroys uterine epithelium and comes into contact with blood vessels, without destroying endothelium[71,72]. The main part of fox placental trophoblast in which active proliferation and polyploidization takes place is out of contact with the glandular zone of endometrium and forms the fetal part of placenta[73]. The giant trophoblast cells are scattered throughout the fetal part of placenta (Figure 7), forming the largest accumulation near absorptional zones.
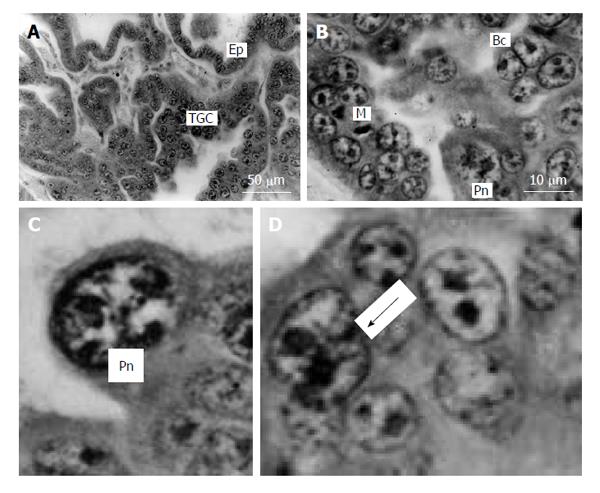
Figure 7 Silver fox placenta.
A and B: Trabeculae of trophoblast and folds of uterine glandular epithelium (Ep) mutually contact each other, trophoblast giant cells (TGC) are scattered in the fetal part of placenta between accumulations of proliferative cells; B: Mitotic (M), binucleate (Bc) and cells with polytene nuclei (Pn); C: Nucleus with non-classic polytene chromosomes; D: A polyploid nucleus in the beginning of fragmentation (arrow). Meyer hematoxylin staining.
DNA content in the trophoblast cells corresponds predominantly to 4c-64c, the main peaks lying at 16 c and 32c; meantime, the highest ploidy corresponded to 64c[73]. Therefore, the ploidy level of the silver fox is lower as compared to the giant trophoblast cells in rodents. Another peculiarity is that there was a considerable deviation from (2n)c, with a tendency to 2n× 3c values and a great variety of the intermediate values suggesting a significant incidence of aneuploidy[73].
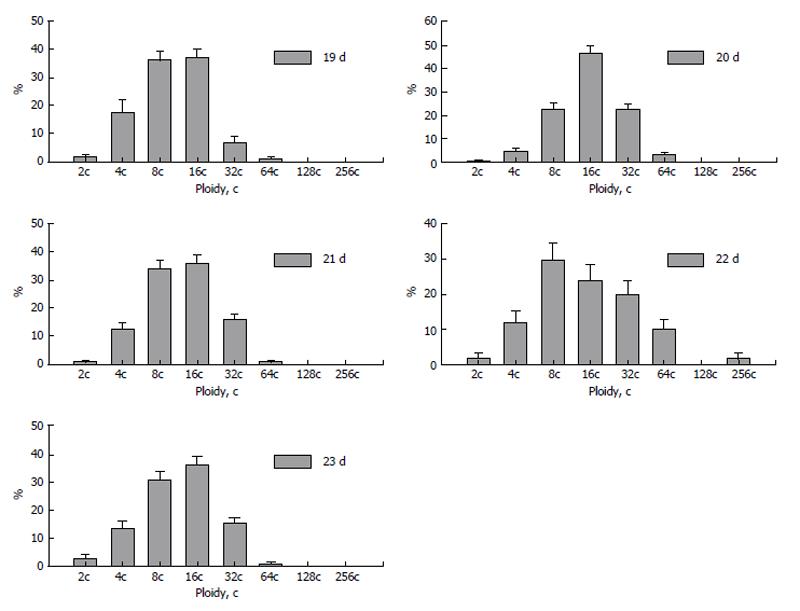
Dynamics of polyploidization during placentation showed several steps of increase and decrease of ploidy levels. At the 20th day of pregnancy the trophoblast nuclei reach the highest ploidy levels (Figure 8). At the next, 21st day, percentage of nuclei of higher ploidy levels decreases. This trend is also seen at the 22nd day: the “divergence” of ploidy is observed, the percentage of diploid and tetraploid nuclei rises to 10% and 20%, respectively; simultaneously the percentage of 32c and 64c nuclei also increases as compared to that at the 21st day, and trophoblast cell population reaches the highest ploidy levels-128c and 256c. At the 23rd day, a new “wave” of polyploidization, similar to 20 d, takes place. The reason of such a fluctuation may be accounted for appearance of new zones of trophoblast cell proliferation. Therefore a part of trophoblast cells divide mitotically providing diploid and low-polyploid cells whereas other cell undergo endocycles to reach high ploidy levels. Simultaneously, pictures similar to nuclear fragmentation described in the rodent giant trophoblast cells (see above) were observed in the fox trophoblast[73]; such a genome segregation also may be a reason for a decrease of the ploidy level.
Figure 8 Dynamics of polyploidization of the trophoblast cells in silver fox placenta.
The DNA content measurement in mitotic figures showed 4c, 8c, and 16c mitotic figures that confirm the ability of 2c-8c to divide mitotically. The presence of binuclear cells, polyploid mitoses, and atypical metaphases and anaphases including multipolar mitoses indicates acytokinetic and uncompleted mitoses as a way by which the cells reach low (4c-8c) ploidy levels[6]. The higher ploidy levels may be attained by switch to endoreduplication cycle; it is confirmed by the presence of nuclei with characteristic non-classic polyteny (Figure 7). Interestingly, histograms of DNA content in the mitotic figures also show a tendency for “triploidy”. It should be noted that signs of aneu- or/and triploidy are intrinsic of fox trophoblast: the uterine epithelial cells that also polyploidize reaching 4c-8c show quite euploid DNA content histograms[73].
The reason for deviation from euploidy in the fox trophoblast cells was not clarified completely. However, it cannot be ruled out that it may result from depolyploidization as a result of the multipolar mitoses and the process similar to the nuclear fragmentation described in the previous chapter.
PECULIARITIES OF HUMAN TROPHOBLAST GENOME MULTIPLICATION
Human trophoblast shows, at the first glance, a great difference in the trophoblast polyploidization as compared to rodents, which probably is accounted for by human placenta formation and growth characteristics. During the most of pregnancy, the trophoblast continuous layer at the border of decidua consists of a layer of syncytiotrophoblast, whereas several local zones of the intrauterine invasion of extravillous trophoblast (EVT) are concentrated at tips of the anchoring villi[74]. Numerous data of caryologic analysis as well as DNA flow cytometry showed the prevalence of the diploid or, sometimes, near-diploid chromosome set in the human trophoblast cells[75-77]. Meantime, the human trophoblast invasion show many analogies in regularities of the genome reproduction to trophoblast of rodent placenta.
Thus, in human, like in rat placenta, the deep intrauterine interstitial and endovascular invasion is accomplished at the complete cessation of DNA replication[5,44,74,78]. Lack of genome replication of the invading trophoblast may prevent the ectopic proliferation of the trophoblast cells both within the uterus and in other parts of the maternal organism.
Polyploidization of the extravillous trophoblast cells also show similarities with rodent placenta. Thus, the proliferative EVT attached to the basal membrane of tip of villi are mostly diploid, but the ploidy increases progressively to 4-8c in process of approach to the border of decidua[7,79]. Thus, like in rat, mouse, and field vole, the highest ploidy is characteristic of the human trophoblast cell layer that borders the semiallogenic maternal tissues.
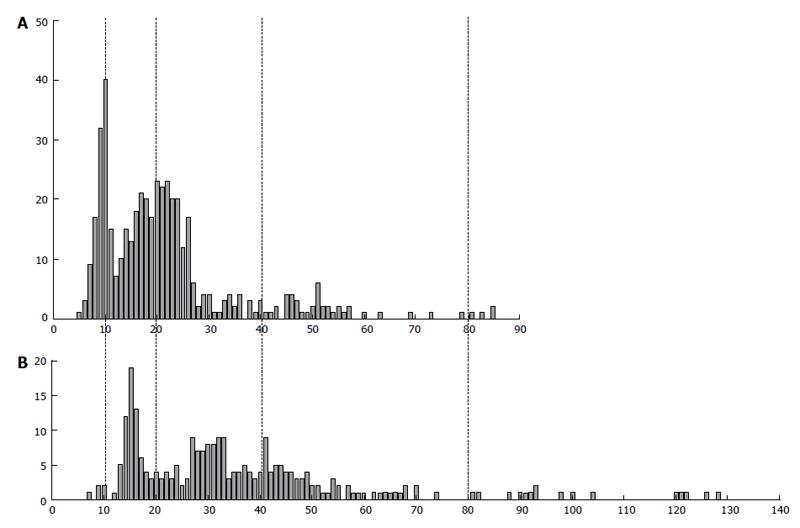
An interesting peculiarity of the human EVT genome reproduction is a ploidy divergence that takes place at the moment of intrauterine invasion (Figure 9)[7,79]. The fraction of 8c cells increased by up to 9.7%; besides, a few 16c nuclei appeared, whereas, on the other hand, the percentage of diploid cells also rise noticeably-on average, from 20% to 30%. Such a tendency was also found in the EVT invaded up to myometrium: the percentage of diploid nuclei exceeded 40%, whereas a number of highly polyploid 8c and 16c nuclei persisted in this zone.
Figure 9 The difference in DNA content in human extravillous trophoblast cells invaded endometrium in placentae of the first trimester between two individuals (A and B).
Abscissa: The DNA content (arbitrary units); Ordinate: The number of cells.
One of peculiarities of the trophoblast cell polyploidization is a great variety of the genome multiplication between individual placentae[79]. Thus, Figure 9 presents histogram of the DNA content distribution in the extravillous trophoblast cells invading decidualized endometrium in placentae of two pregnant women. We can see that one of them shows more or less clear-cut peaks corresponding to 2c and 4c; the latter ploidy class prevails, some cells being able to reach as many as 16c. Another sample shows more extended histogram, in which the main peaks tend to 3c and 6c, the greatest number of cells being located between 4c and 8c. Besides, in the second sample, the noticeable number of cells exceeded 16c. A correlation between DNA content values not multiple to (2n)c and the highest ploidy level is seen here; besides, a tendency for 3c × 2n values is characteristic of the samples of the higher ploidy level.
Endoreduplication of human cervical trophoblast with prevalence of tetraploid cells was also found by means of caryological analysis with fluorescent in situ hybridization (FISH) staining of the chromosomes X, Y, and 21[80].
Endocycle progression in the human EVT is confirmed by downregulation of mitotic cyclins A and B1 alongside the invasion pathway and peculiar expression of Cyclins D and E as well as p57kip2[7].
According to our data, a great number of DNA content values not multiple to (2n)c is observed in the beginning of the EVT invasion. When the cells penetrate endometrium and myometrium, more clear-cut peaks tending to (2n)c are observed[7,79].
Interestingly, some data suggest the necessity of aneuploidy for normal EVT trophoblast invasion. Thus, molecular cytogenetic data showing that approximately 20% to 60% of interphase EVT invasive cells in the normal pregnancies acquired aneusomies involving chromosomes X, Y, or 16[81]. The incidence of aneuploidy positively correlated with gestational age and differentiation to the invasive phenotype. Scoring 12 chromosomes in flow-sorted cytotrophoblasts showed that more than 95% of the cells were hyperdiploid. Thus, aneuploidy appears to be an important component of normal placentation, perhaps limiting the proliferative and invasive potential of cytotrophoblasts within the uterus[81].
A series of recent investigations allow some authors to put forward a concept that transition from endoreduplication to polyploidy and then to aneuploidy represents a genetically diverse pool of cells[18-20]. The authors suggest that a set of these genetically changed cells may be useful and amplified under conditions of stress[18]. In the case of invading trophoblast we can assume that aneuploidy that accompanies the complicated processes of the trophoblast genome reproduction may rise several genotypes that may promote cells to survive under the stress conditions inside semiallogenic maternal tissues. It may be suggested that the most stressful condition for EVT cells is a moment of overriding the border with decidualized endometrium; at this moment the optimal genotypes are selected; later on, deviation of the euploidy decreases. It cannot also be ruled out that simultaneously the cells of higher ploidy are selected to invade the proximal part of endometrium and the small low-ploidy elongated cell to invade deep up to myometrium (Figure 9).
It is not easy to explain the possible significance of tendency to triploidy in the invading human EVT (that may be analogous to the same tendency observed in silver fox). Triploidy of human trophoblast was reported in several papers[76,82]. Very often, it is observed under pathological conditions connected with ectopic or disturbed trophoblast invasion, for example, in the hydatidiform mole[76] or severe preeclampsia[82]. Meantime, basing on the above-mentioned data on the possibility of non-mitotic genome segregation (nuclear fragmentation) as a regular step of trophoblast cell lifespan, we can suggest that such a process may result in separation of the chromosome set non-multiple to (2n)c. This process, most probably, does not include the way of exact distribution of all chromosomes into the daughter cells characteristic of mitosis, that is why it may be suggested that such a way of depolyploidization of trophoblast cells may result in aneuploid genotypes.
The reason for such a variability may be accounted for by the ways of human EVT polyploidization. Endomitosis and non-classic polyteny were observed in the trophoblast cells in normal human pregnancies[32,37,83] as well as in hydatidiform moles[32].
According to our data, the processes like endoreduplication or/and endomitosis, most probably, prevail in the EVT invasive pathway. The Figure 10 shows a chorionic villus tip stained with DAPI. A great number of cells with numerous chromocenters as well as nuclei at different stage of endomitosis are present there (Figure 10)[38]. Meantime, a number of nuclei with enlarged chromocenters, whose number was not increased with increasing ploidy, suggest a possibility of passages of several rounds of polytenization[38]. Thus, it cannot be ruled out that a relatively low ploidy level in the human placenta prone to aneuploidization may be linked with endoreduplication/endomitosis that, theoretically, may be involved in single/double/triple genome segregation processes.
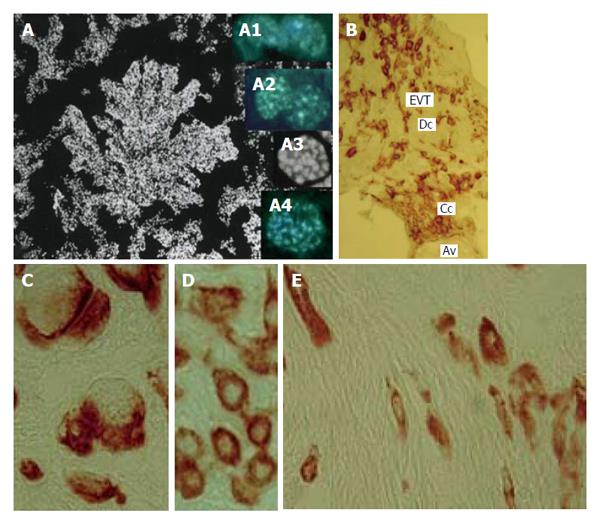
Figure 10 Human trophoblast cells undergoing polyploidization.
A: A squash spread of chorionic villus stained with 4’,6-diamidino-2-phenylindole; A1: Binucleate cell in interphase; A2: Binucleate cell in prophase; A3: Endometaphase; A4: Endoanaphase; B: A cell column (Cc) at the tip of anchoring villus (Av) generates a pool of extravillous trophoblast cells (EVT) capable for invasion of decidualized endometrium (Dc); C: Multinucleate EVT invaded decidua; D: Interstitial EVT of moderate ploidy; E: Small elongated low-ploidy EVT that reach myometrium.
GENOME MULTIPLICATION IN THE TROPHOBLAST CELLS OF RUMINANTS
Ruminants represent a mammalian group with the so-called epitheliochorial placenta, in which trophoblast invasiveness is minimal: the trophoblast cells attach to the uterine epithelium mostly without its degradation[84]; meantime, the trophoblast cells express the same integrins as the highly invasive human trophoblast cells[85]. The bovine trophoblast cells that come into close contact to the uterine epithelium retain their mitotic activity and reach the ploidy level 4c-8c via reduced (restitutional) mitoses[11,30]. Tripolar mitoses suggesting a possibility to reverse to the lower ploidy level also are present there. It is quite probably that lack of deep invasion that does not imply phagocytosis of epithelium, blood or decidual cells by the trophoblast does not require switch to the endoreduplication cycle, although a low level of ploidy is still necessary for such a mode of feto-maternal interaction.
The most striking example of polyploid mitoses was described in alpaca trophoblast. Elevated nuclear DNA contents in the giant trophoblast cells of alpaca could be achieved by modified cell cycles with a complete lack of mitosis (endoreduplication) or with incomplete mitoses[12]. Electron microscope observation made on serial sections revealed that TGCs are truly multinucleate with several highly lobulated nuclei. Feulgen staining showed that TGC nuclei have the higher DNA content than nuclei of other trophoblast cells. The number of argyrophilic nucleolar organizer regions in nuclear profiles of TGC was between 15 and 100; numerous nucleoli suggest polyploidization, in which mitoses take part, as this was observed in the rat decidual cells[86]. In the latter case, numerous decidual cells are formed by active mitotic divisions; numerous binucleate cells were observed, there were many enlarged mitoses of irregular form, a part of mitoses were tri- and tetrapolar. Very often, bi-tri- and tetranuclear cells were observed that could result from multipolar mitoses, the nuclear ploidy reached 8c. Numerous nucleoli (up to 20) were also observed in the lobulated nuclei. As to alpaca trophoblast, even larger multipolar mitotic figures with maximal diameters of 80 μm were observed in placentas on gestation days 264 and 347. No cytokinesis was seen in TGC[12]. The authors note that subsequent acytokinetic mitoses may lead to accumulation of chromosomes and centrioles in TGC. With increasing ploidy levels, the shape of these polyploidizing mitoses becomes more irregular. The restitution of nuclei after these complex multipolar mitoses is likely to result in the irregular nuclear shape in TGC. Therefore, it is an exceptional example of polyploidization via restitutional (reduced) mitoses that probably may lead to the high ploidy level. It seems doubtful that in this case the multipolar mitoses may result in depolyploidization.
Thus, a conclusion can be made that shallow-invasive trophoblast cells of ruminants involve polyploidization via restitutional (reduced) mitoses.
CONCLUSION
The data considered here demonstrate that trophoblast cells of different mammalian species are characterized by different modes of multiplication of their genome that, probably, is linked with their ploidy level, capability for further proliferation, necessity of irreversible or, on the contrary, of reversible polyploidization that, in turn, most probably, is accounted for by the trophoblast cell specific function.
One of the most important advantages of polyploidy for the trophoblast cells contacting semiallogenic maternal tissues may be the delay of proliferation to avoid segregation of the damaged chromosomes[19]. Besides, multifold genome doubling makes the endocycling cells more resistant to mutagens. Thus, the highly endopolyploid murine TGC are much more resistant to irradiation than the low-ploid trophoblast cells[42]. Interestingly, endoreduplication as a response to mutagens can be induced experimentally, and some regulatory pathways were recently revealed. Thus, following double strain breaks induction in the root tips of Arabidopsis by Italic the cells switch to endoreduplication[87]. This cell alteration requires the plant-specific transcription factor Suppressor gamma response 1 which transmits signals from the conserved Ataxia Telangiectasia mutated and Ataxia Telangiectasia-mutated and RAD3-related DNA damage sensor kinase[86]. This DNA break response produces transcriptional changes that are consistent with downregulation of mitotic factors and upregulation of cell cycle genes that promote endoreduplication.
Recently there were obtained some other confirmations of significance of non-mitotic polyploidization under condition of DNA damage. In Drosophila by Italic, endoreduplication cells acquire resistance to DNA damage through a mechanism involving the silencing of cell death genes[19,88]. Similarly, endoreduplication mouse trophoblast cells that undergo endoreduplication also downregulate the DNA damage response. During differentiation of trophoblast stem cells into polyploid TGC, the protein level of damage-responsive Chk1 is decreased providing for endoreduplication. This decrease in Chk1 enables polyploid trophoblast cells to evade apoptosis through suppression of the DNA damage pathway[89,90].
It seems to be obvious that there are two main ways of genome multiplication: endoreduplication that involves downregulation of mitotic events and reduced mitosis (“endomitosis”) in which entrance into mitosis is a prerequisite of genome multiplication. Endoreduplication allows to combine growth and specific functioning of cells that retain their peculiar organization. On the contrary, the cells that polyploidize via reduced mitoses retain their mitotic potential necessary, for example, for accumulation of a great number of cells, or for regeneration. In case of rodent trophoblast, the highly polyploid TGC undergoing endoreduplication serve a barrier between semiallogenic tissues whereas highly proliferative low-ploid trophoblast cell populations give rise to numerous JZ and labyrinth trophoblast cells.
Endoreduplication is a characteristic of highly invasive trophoblast cells. It is confirmed by comparison of placentation of mammalian species. Thus, highest invasiveness is characteristic of rodent and human placenta in which invasive trophoblast cells undergo endoreduplication whereas reduced mitoses are observed in the low invasive epitheliochorial placenta of ruminants.
Another advantage of polyploidization that is widely discussed now is a possibility to gain a variety of genome changes. Such a possibility was recently demonstrated in hepatocytes[17,18,21]. Apart from polyploidization by acytokinetic and subsequent normal (polyploidizing) mitoses, hepatocytes may undergo depolyploidization that result from multipolar mitoses[18]. Indeed, during multipolar mitosis, microtubules from different poles of spindle can be attached to a single kinetochore, and failure to repair such merotelic attachment can lead to incomplete chromosome segregation. In this case, chromosome bridges and laggings were observed. Karyotype analysis showed high frequency of aneuploidy in the normal murine liver: nearly 25% of hepatocytes from the 3-wk old mice were aneuploid, the aneuploidy increasing to 70% in the 4-15-mo old mice[18]. Interestingly, the entire chromosomes were gained and lost in this case, and structural rearrangement was rarely seen; besides, all chromosomes of genome were affected equally. One or more chromosomes were gained or lost by each aneuploid hepatocytes, and occasionally chromosome gains balanced losses mimicking the normal chromosome number. That is why, probably, aneuploidy did not lead to noticeable deviation from (2n)c seen in histograms of the DNA content distribution in hepatocytes measured by DNA cytometry[91]. The karyotype and FISH analysis also revealed a significant level (30%-90%) of aneuploidy in the human liver[17]. Strikingly, gain and loss of chromosomes in hepatocytes under stress conditions may result in selection of the specific karyotype that can result in adaptation to injury[20].
The statement that polyploidy is often accompanied by aneuploidy may be accounted for by the fact that aneuploidy often results from depolyploidization that, in turn, requires polyploid cell formation. Meantime, it should be kept in mind that genome multiplication, in many cases, is irreversible. In this connection, a spectacular example is given by the ovarian follicle nurse cells[42]. When the cells reach 32c, centrioli leave their position near nucleus and move into oocyte through the cytoplasmic bridges. This is a way by which the cell “burns the bridges” to ploidy reversal and renewal of mitotic divisions. In the majority of cells normally undergoing endocycles, the genome multiplication is irreversible[22]. Therefore, aneuploidy may arise in the modified cell cycles prone to depolyploidization. Actually, at present little is known as to which cell types may be capable for the spontaneous non-pathologic aneuploidization.
Endopolyploidization as an escape route from cell death has been investigated on cancer cells. It has been demonstrated that after irradiation the great majority of cells die due to a mitotic catastrophe[92]. However, some cells escape the mitotic catastrophe and polyploidize. A few endopolyploid cells undergo depolyploidization and create a set of para-diploid viable cells capable for mitoses that may give rise to subclones[92,93]. Recent data of this research group indicate that tumor cells can induce opposing processes of senescence and stem cell generation in response to these treatments whose biological significance and molecular regulation currently are poorly understood[94]. Although cellular senescence is typically considered a terminal cell fate, it was recently shown to be reversible in a small population of polyploid cancer cells induced after DNA damage. Overcoming genotoxic insults are associated with reversible polyploidy. The subsequent depolyploidization results in evoking the self-renewal potential in survived cells[94].
In rodent placenta, the secondary TGC provide an example of irreversible differentiation that, however, results in formation of the low-ploidy subcellular compartments that may behave like viable cells except for renewal of their capability for proliferation. Theoretically, depolyploidization may take place in the JZ and labyrinth trophoblast cells of rat and field vole undergoing the polyploidizing mitoses, some amount of multipolar mitoses being observed here[6]. Deviation from (2n)c in human and silver fox trophoblast[7,73,79] suggests a possibility of aneuploidy and other chromosome changes (aberrations, etc.) that may be even more strongly pronounced in trophoblast cells of these species. It cannot be ruled out that in human trophoblast a high degree of genomic changes also may be important for selection of cells able to survive under conditions of deep intrauterine invasion. In silver fox, a series of endoreduplication and proliferation cycles involving depolyploidization via multipolar mitoses and a process similar to nuclear fragmentation would also form a multifunctional system resistant to injury from the maternal tissues.
It seems to be important that different mammalian species have different programs of genome multiplication. Thus, placenta of rodents with strict spatial location of high- and low-ploidy trophoblast cells, with low incidence of deviation from (2n)c, probably is accounted for by their strict developmental program allowing them to produce their progeny during a limited period of short pregnancy. The mammalian species with a long period of pregnancy probably stick to another strategy allowing them to generate a more specific response to stress factors that may appear occasionally during months of intrauterine development. In these cases, a more diverse genome changes followed by selection of favorable genotypes may be useful to maintenance of placental functions.
ACKNOWLEDGMENTS
The authors are grateful to Leonid Z Pevzner, MD, PhD, the chief interpreter of Institute of Cytology of the Russian Academy of Sciences for his help in translation and edition of the manuscript.