Published online May 27, 2014. doi: 10.5496/wjmg.v4.i2.6
Revised: January 23, 2014
Accepted: April 3, 2014
Published online: May 27, 2014
Processing time: 168 Days and 23.1 Hours
The acquisition of a storage information system beyond the nucleotide sequence has been a crucial issue for the propagation and dispersion of RNA viruses. This system is composed by highly conserved, complex structural units in the genomic RNA, termed functional RNA domains. These elements interact with other regions of the viral genome and/or proteins to direct viral translation, replication and encapsidation. The genomic RNA of the hepatitis C virus (HCV) is a good model for investigating about conserved structural units. It contains functional domains, defined by highly conserved structural RNA motifs, mostly located in the 5’-untranslatable regions (5’UTRs) and 3’UTR, but also occupying long stretches of the coding sequence. Viral translation initiation is mediated by an internal ribosome entry site located at the 5’ terminus of the viral genome and regulated by distal functional RNA domains placed at the 3’ end. Subsequent RNA replication strongly depends on the 3’UTR folding and is also influenced by the 5’ end of the HCV RNA. Further increase in the genome copy number unleashes the formation of homodimers by direct interaction of two genomic RNA molecules, which are finally packed and released to the extracellular medium. All these processes, as well as transitions between them, are controlled by structural RNA elements that establish a complex, direct and long-distance RNA-RNA interaction network. This review summarizes current knowledge about functional RNA domains within the HCV RNA genome and provides an overview of the control exerted by direct, long-range RNA-RNA contacts for the execution of the viral cycle.
Core tip: This review summarizes the main aspects of structurally conserved genomic RNA elements in the hepatitis C virus (HCV) genome and their role in the viral cycle. The genome of RNA viruses is a dynamic genetic entity endorsed with an information storage system defined by highly conserved, complex structural units, termed functional RNA domains. The genome of HCV contains several well-studied functional RNA domains that control essential viral processes, as well as transitions between them, by recruiting protein factors and also by establishing a complex, direct and long-range RNA-RNA interaction network.
- Citation: Romero-López C, Berzal-Herranz A. Structure-function relationship in viral RNA genomes: The case of hepatitis C virus. World J Med Genet 2014; 4(2): 6-18
- URL: https://www.wjgnet.com/2220-3184/full/v4/i2/6.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v4.i2.6
The genomes of RNA viruses are not passive elements. The inherent high error rate of the viral polymerase during replication provides an important evolutive advantage by the generation of genotypically and phenotypically different virus pools on which natural selection operates[1]. By using this strategy, viruses have got RNA genomes with numerous signals overlapping protein coding sequences, thus achieving multiple levels of regulation throughout the infectious cycle. All this information is compactly packed in a minimal size for optimal propagation. Viral RNA genomes use a information storage system beyond the nucleotide sequence, defined by highly conserved regions that exhibit complex folding and play direct, functional roles in the viral cycle[2-4]. Two levels of structure or folding can be distinguished within an RNA molecule: (1) the secondary structure involves double and single stranded regions arrangements; and (2) the tertiary structure is determined by the relationships established between secondary structure elements. The combination of both conformational levels establishes the final shape of the RNA to generate the so-called functional RNA domains. These are dynamic elements since their structure can be selectively adopted from a wide variety of possible foldings to execute a specific function by recruiting protein factors, or modulating the conformation and function of distant regulatory elements[5]. These mechanisms achieve an active control of the gene expression. Therefore, RNA folding acts as a regulatory machine to diversify RNA genome functions with a minimal size.
Functional RNA domains are typically identified as one or more stem-loops with highly conserved sequence motifs located in the loops. These elements were initially described located in the 5’-untranslatable regions (5’UTRs) and the 3’UTRs of viral genomes, but now evidences are accumulating for their widespread distribution throughout the entire genomic RNA[5]. They can be organized, either as well-defined, phylogenetically conserved RNA structural motifs, or as sets of extensive folded regions throughout the whole viral genome [genome-scale ordered RNA structures (GORS)], following a clear structural pattern that may change even between closely related viruses[6,7].
The recent advent of novel bioinformatic tools and experimental techniques to probe and study RNA structure has provided high-resolution pictures of numerous viral RNA molecules. Among them, structural elements of the hepatitis C virus (HCV) genomic RNA are one of the best characterized from many different viruses. HCV infection affects to more than 3% of the world population, with high incidence of cirrhosis, hepatic steatosis and hepatocellular carcinoma. To date, no efficient vaccines have been developed against HCV and current treatments based on pegylated-interferon α and ribavirin are the standard of care (SOC) regimen with a limited efficacy of around 40% of the patients. Additionally, this therapy has important side effects. Recently, two direct-acting antiviral drugs targeting the viral protease NS3, telaprevir and boceprevir, have been approved by the United States Food and Drug Administration[8]. These compounds can be administered in conjunction with pegylated-interferon α and ribavirin for a short period of time to achieve an improved sustained virological response[8] with respect to the SOC. Unfortunately, prolonged treatments lead to the appearance of resistant variants. Other drugs targeting either the protease NS3 (simeprevir) or the viral polymerase NS5B (sofosbuvir) are currently being tested in Phase II/III clinical trials.
HCV belongs to the Flaviviridae family, which includes yellow fever virus, bovine diarrhea virus and dengue virus. The HCV genome shows such a variability that up to six different genotypes, with hundreds of subtypes and isolates, have been identified[9,10]. Viral genotype clearly affects the success of interferon therapy, although no clear correlation with virulence exists. Further, the HCV population infecting a patient is structured in terms of quasispecies. This term defines the closely related sequences of a heterogeneous viral population infecting a single individual[11]. Quasispecies structure has been associated with the failure of infected people to clear the virus and the subsequent development of a chronic infection[12]. Therefore, the identification of conserved therapeutic targets and the search for fully effective antiviral compounds is a major goal of HCV research. The functional importance of genomic structural elements for virus persistence and their high conservation rate suggests they might make good therapeutic targets. This review focuses in the main structural features of the HCV genomic RNA functional domains and their roles in the viral cycle.
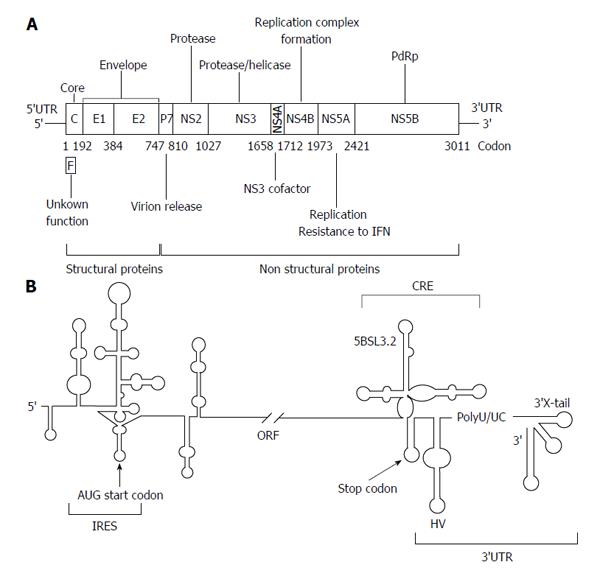
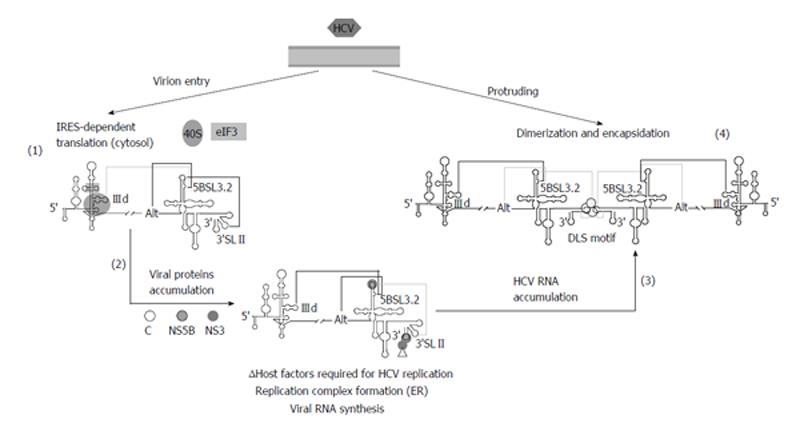
The HCV genome is about 9600 nucleotides-long, single stranded positive RNA molecule[13-15] that encodes for a single open reading frame (ORF) flanked by two highly conserved UTRs (5’UTR and 3’UTR) (Figure 1A). The viral genome controls important processes of the infective cycle. During early infection, the genome acts as mRNA to generate the viral structural (core protein C, p7 and the envelope proteins E1 and E2) and non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B). HCV translation is initiated by an internal ribosome entry site (IRES)-dependent mechanism[16,17] different to the cap-dependent method used for most cellular mRNAs. The IRES element is mostly located at the 5’UTR and spans a short stretch of the core coding sequence[18,19] (Figure 1B). Both the initiation translation step and the subsequent elongation phase are influenced by the presence of domains located at the 3’ end of the HCV genome[20-25]. This process is dependent on the acquisition of a circular topology resembling the closed-loop structure adopted by cellular cap-mRNAs. Such architecture is achieved by both the recruitment of protein factors, able to simultaneously bind to the 5’UTRs and 3’UTRs of the genomic HCV RNA[20-23,26-28], and also by the establishment of direct, long-range RNA-RNA interactions[29-31]. Once viral proteins levels have reached a certain threshold, the genomic RNA serves as a template to initiate replication at the 3’UTR in a structure dependent manner. This process is also influenced by the 5’ end of the HCV RNA[32,33]. The accumulation of viral genomes enhances the formation of homodimers by the interaction of two viral RNA molecules in the presence of the core chaperone protein[34-37]. Packaged genomic RNA is finally enveloped and released to the extracellular environment.
The maintenance of a proper balance between these processes involves fine regulation mechanisms, which involve the interplay of functional RNA domains located throughout the entire ORF[38,39]. The 5’ core coding sequence helps in the preservation of structures important for IRES activity and replication (Figure 1B)[19,40-43]. Within the 3’ end of the NS5B coding sequence, the stem-loop 5BSL3.2 is embedded in a cruciform structure that has been identified as a cis-essential element for viral RNA synthesis [cis-acting replicating element (CRE)] (Figure 1B)[44,45] and as a regulatory partner of the IRES function[25].
An interesting feature of all these functional RNA domains is that they do not operate only by recruiting protein factors. Instead, they establish a complex and dynamic network of contacts, which fits viral necessities to promote the consecution of different steps of the viral cycle, as well as the switch between them. Furthermore, this interacting web provides important benefits, such as minimizing protein requisites.
Next sections will outline the current knowledge about different HCV functional RNA domains and their involvement in the complex interaction network that governs the initiation of essential viral events and the transitions between them.
The initiation of the HCV protein synthesis is driven by the high affinity interaction IRES-40S[46-48]. This primary contact promotes conformational changes that directly clamp the viral RNA to the ribosomal subunit and thus position the appropriate start codon in the P site[49]. The further binding of eIF3 aids the incorporation of the ternary complex eIF2-GTP-tRNAiMet to yield the 48S particle[48,50]. The formation of the active translation complex 80S is assessed by the GTP hydrolysis for the concurrent release of eIF2 and eIF3[51] and the final joining of the 60S subunit. It is noteworthy that this mechanism is primarily accomplished by functional RNA domains, thus minimizing protein factor requirements and simplifying the pathway for the assembly of the fully active ribosome.
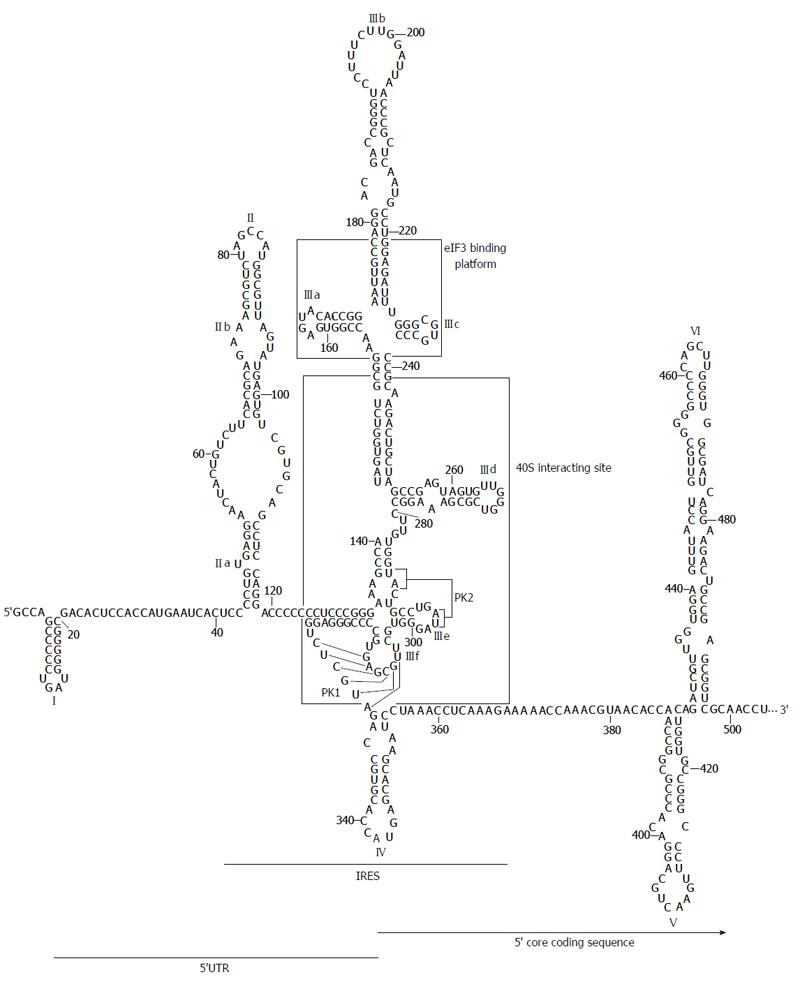
The secondary structure of the HCV IRES region was originally proposed by Brown et al[52] and latter refined to include several new motifs and interactions. Under physiological magnesium conditions, the HCV IRES folds into two major domains with well defined functions (II and III; Figure 2)[53], plus a short stem-loop containing the start codon (domain IV)[54]. Rather than forming a tightly packed element, domains II and III are extended and aligned at both sides of a complex double pseudoknot structure (PK1 and PK2; Figure 2)[49,55]. The 3D architecture and several single RNA structural elements are highly conserved among other closely related viruses from the Flaviviridae family[46,56,57].
Domain II is an autonomously folded module composed of two short helical segments, the basal subdomain IIa and the apical subdomain IIb, separated by a highly conserved internal E-loop[56,58] and capped by an apical loop (Figure 2). Domain II adopts an overall distorted L-shape conformation[59] because of the twist forced by the internal E-loop. This folding is conserved in HCV and related viruses[51].
While domain II is not essential for 40S recruitment[47,60,61], it has been shown that its deletion decreases viral protein synthesis yield up to five-fold by blocking the formation of the translationally active 80S complex[46,48,60,62,63]. Analysis by cryo-EM have demonstrated that the bend in domain II is a requisite for changing the conformation of the 40S ribosomal subunit[49,64], in a reminiscent manner to that shown by eIF1 in the canonical cap-dependent translation initiation mechanism[65]. The apical loop placed in subdomain IIb would also contribute to this structural rearrangement in the ribosome[66]. Remarkably, all these conformational reorganization events do not only account on ribosomal proteins but also on the 18S rRNA. This could be the result of the coordinated action mediated by long-distant contacts established between domains II and IV[67,68]. Ribosome folding rearrangements further induce eIF2-GTP hydrolysis, triggering the release of protein factors and the recruitment of the 60S subunit to constitute the 80S complex[48,51,63,64].
The large, highly branched domain III consists of six hairpins (designated subdomains IIIa to IIIf) organized around three- and four-way junctions (Figure 2)[52], which can be identified as recruiting centers for the translational machinery. The apical IIIabc junction is the platform for the binding of eIF3[50,69]. The main goal of this interaction seems to be the relief of the competition between eIF3 and the IRES for a common site in the 40S ribosomal subunit, as well as avoiding the formation of canonical 43S translational complexes[70]. This assesses that HCV mRNA translation is specially favored over that of host mRNAs.
The middle section of domain III is defined by a three-way junction that contains the essential G-rich subdomain IIId (Figure 2). This element is the core 40S binding center[60,71,72]. Its structure is that of a dynamic stem-loop with an internal E loop motif and an apical loop with typical U-turn geometry[73,74]. This architecture exposes the bases placed in the apical loop and favors their interaction with viral and host ligands, both nucleic acids and proteins. Further, the subdomain IIId seems to be a determinant partner in the acquisition of the functional folding of the surrounding domains[53].
The basal fragment of domain III (subdomains IIIe and IIIf) includes the highly conserved, complex double-pseudoknot motif (PK1 and PK2; Figure 2)[55], which defines a four-way junction to constrain the position of the AUG codon at the P-site of the 40S ribosomal subunit. Remarkably, the spatial distance between the pseudoknot and the AUG firmly resembles to that observed between the canonical Shine-Dalgarno motif and the initiation codon in prokaryotic mRNAs[75]. As noted above, the structural element PK1-PK2 also guides domains II and IIIabc in an extended conformation to get the easy access of protein factors.
Domain IV exposes the AUG start codon, at nucleotide 342, in an apical loop enclosing a helical motif (Figure 2). This structure is not conserved in other HCV-like IRESs[76]. In fact, the stem must be unwound to allow for the recognition of the AUG codon, which could entail some disadvantages. This is in good agreement with data demonstrating that the stability of stem-loop IV is inversely correlated to IRES translational efficiency[54].
Therefore, the HCV IRES is defined by a set of RNA domains that replace the functions played by many host factors to provide a simplified way for the initiation of the viral proteins synthesis. Moreover, these functional domains are able to manipulate the translational machinery to assess the preferential reading of the HCV mRNA.
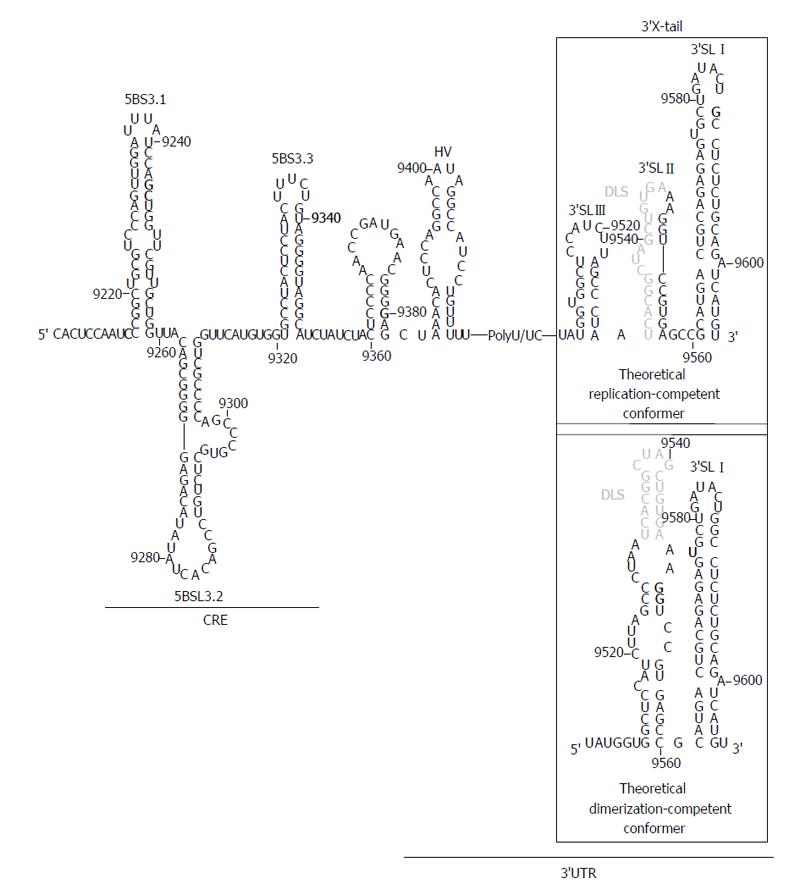
The HCV 3’UTR is of primary importance for the initiation of the minus RNA strand synthesis during the viral replication step[33,77,78] and also may act as enhancer of the IRES function[20-24]. It is about 240 nts long sequence placed at the 3’ end of the viral genome[79], with evolutionarily conserved secondary structure elements that define three functionally and conformationally independent modules (Figure 3). From 5’ to 3’: (1) A poorly conserved sequence of around 40 nts, termed hypervariable region at the 5’ end of the 3’UTR. It folds as a single stem-loop, which is not completely required for viral replication[33,78]; (2) A polyU/UC tract, whose length and composition is a critical determinant of efficient HCV replication in cell culture[80]. It has been proved that a minimum of 26 U nts homopolymer is enough for efficient amplification of the viral RNA[33,78]. Further, it can act outside of its usual molecular context, thus suggesting that this is not only a linker region[80]. The polyU/UC stretch also interacts with host factors related to cellular protein synthesis, such as polypyrimidine tract-binding protein[81,82], the La autoantigen[83], heterogeneous nuclear ribonucleoprotein C (Gontarek, 1999 #1925) and glyceraldehyde-3-phosphate dehydrogenase[84], among others[85,86]. It seems likely that the recruitment of these factors could contribute to regulate viral translation mediated by the IRES region[20,21]; and (3) The 3’X-tail is a highly conserved, 98-nts long sequence, located at the 3’ termini of the HCV genome. It theoretically folds into two alternate and mutually exclusive conformations[35] (Figure 3). Both predicted structures preserve the essential 3’SLI placed at the very 3’ end, which has important implications for the initiation and specificity of the viral RNA replication[87-89]. The 55 nts segment placed upstream of 3’SLI folds either as two stem-loops, named 3’SLII and 3’SLIII, or as a single stem-loop exposing a 16 nts long palindromic sequence [dimer linkage sequence (DLS)] (Figure 3). Both conformers assume different functionalities during the HCV cycle and are therefore related to transitions between different steps of the viral infection.
The molecular basis of the 3’UTR functioning are not well understood. Several reports have described the binding of both viral and host factors to the different structural elements of the 3’UTR[81,90-97], but these findings do not provide completely satisfactory explanations for many of the experimental observations. The involvement of the 3’UTR in a long range RNA-RNA interaction network with other genomic elements would likely fill the gaps in the complex functioning of this region[30,31,33,98,99].
Advances in novel bioinformatic tools have allowed for the extensive search of evolutionarily conserved RNA domains, resulting in the identification of domains distinct from those present in the UTRs. Comparative analyses of numerous HCV isolates sequences revealed an unusual high degree of conservation in the 5’ end of the core protein coding sequence[100]. Interestingly, this conservation could not be explained only by the preservation of the amino acid sequence since synonymous substitutions were suppressed. This finding entails a functional constrain that was related to the presence of an alternative ORF coding for the so-called protein F[101,102] (Figure 1A); and to the existence of structural RNA domains with functional roles in the HCV cycle[38] (Figure 2). While the production and biological role of protein F is still a controversial issue[103], it has been demonstrated that the 5’ core coding sequence folds as two stem-loops (domains V and VI; Figure 2) important for IRES activity and viral replication[19,40,42,43].
The mechanism of action of domains V and VI is unclear. It has been proposed their participation in a long-range RNA-RNA interaction involving nucleotides 24-38 of the linker region between domains I and II in the 5’UTR, and 428-442 placed in domain VI (Figure 4)[40,41]. This contact would render a locked conformation of the IRES, which could be released by the interaction of the liver-specific microRNA miR-122 with nucleotides 22-28 of the HCV RNA[104]. This hypothesis provides a mechanism for the involvement of domain VI in viral translation, as well as supporting the essential role of miR-122 in HCV infection[105,106]. Alternatively, Roberts et al[107] found that viral translation regulation mediated by miR-122 is strictly dependent on Argonaute proteins and does not involve the structural transition previously proposed. It should be noted that investigations were performed with different experimental tools and model systems. Hence, it is not possible to discard any of the proposals; neither they are mutually exclusive in a cellular context.
In addition to the core coding sequence, the 3’ end of the HCV ORF also harbors evolutionarily conserved structural RNA elements. Up to six different stem-loop motifs have been identified by using a combination of sequence alignment and thermodynamic folding softwares, as well as classical comparative analysis[38,39,108,109]. One of these structural elements, the so-called domain 5BSL3.2 or SL9266, is embedded into a cruciform structure delimited by two adjacent stem-loops, 5BSL3.1 and 5BSL3.3 (CRE, Figure 3). While the essentiality of 5BSL3.2 for virus replication has been largely demonstrated[44,45,98,109], the role of the two additional domains 5BSL3.1 and 3.3 is still unclear[80].
The 5BSL3.2 stem-loop consists of two G-C rich helices connected by an eight-base bulge, and capped by a 12-base apical loop (Figure 3)[45,98]. Disruptions in either the sequence or its folding lead to replication-incompetent HCV genomes[45,98]. Moreover, subtle changes in the apical loop prevent RNA replication, indicating that sequence specificity is required for interaction with protein factors, such as the NS5B protein (viral RNA dependent RNA polymerase)[110] and, more likely, distal RNA functional elements[29,98,99,109]. Relocation of 5BSL3.2 was only possible to the 3’ variable region preceding the poly(U/UC) tract, involving a functional link with the 3’UTR[98]. Domain 5BSL3.2 has been also shown to act as an inhibitory element of the viral IRES function[25], even in the presence of a translational enhancer such as the HCV 3’UTR. This action is strictly dependent on the sequence and the structural integrity of the bulge, pointing again to the existence of interactions with distant functional RNA domains of the viral genome.
As it has been mentioned, the preservation of a proper equilibrium among different viral process and the adequate transitions between them must be assessed for reaching adaptive fitness and virus persistence. To accomplish this, the available functional genomic RNA domains establish an intricate and dynamic interacting web that is mediated, not only by the well-known protein-related 5’UTR-3’UTR bridges[21-23,27,28,111], but more importantly by the formation of direct RNA-RNA contacts that minimize protein requisites.
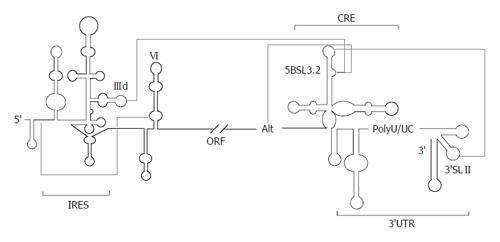
The domain 5BSL3.2 is a good example of an all-RNA-based mechanism. This element participates in viral translation and replication by its integration in a complex network of interactions with distant regions of the HCV genomic RNA (Figure 4). The apical loop of 5BSL3.2 is complementary to the apical loop of the 3’SLII within the 3’X-tail[98,99,112]. The resulting kissing loop contact contributes to the structural organization of the 3’X-tail and is essential for HCV replication[98]. The 8-nts bulge may establish two different interactions: (1) one with the apical loop of the subdomain IIId of the IRES region[29,112], which is related to the aforementioned translational inhibitory effect[25]; (2) the second with the Alt sequence, centred around position 9110, upstream of the CRE element[99,109,112]. This interaction is again critical for the synthesis of the viral genomic RNA[109]. Analyses by different biochemical techniques have proved that the complex interplay IRES-5BSL3.2-3’X-tail influences the global architecture of the affected regions and the surrounding functional RNA elements[30,31,99]. Thus, the 3’ end of the HCV RNA genome, which contains both the CRE and the 3’UTR elements, fine-tunes the three dimensional structure of the IRES region[30], which could be associated to the regulation of viral translation[25]. Conversely, the interaction IIId-5BSL3.2 induce structural rearrangements in the 3’X-tail that finally lead to the conformational transition of the essential domains 3’SLII and 3’SLIII, which switch to a single stem-loop folding that exposes the DLS motif in an apical loop[31] (Figure 3). Importantly, it has been recently reported that all these interactions are equally probable[112]. Therefore, choosing between different contacts might depend on the presence of additional host and/or viral proteins.
Based on these findings, it has been recently proposed a working model[31], which integrates current knowledge concerning to RNA-RNA interactions in the HCV genome, and their implications for the consecution of the viral cycle (Figure 5). In the first stage of the infection, the HCV IRES would be occupied by the translational machinery, thus avoiding any contact with the 5BSL3.2 domain. This would favor the establishment of the interactions 5BSL3.2-3’SLII, which occludes the DLS motif, and 5BSL3.2-Alt. After protein synthesis, the CRE and the 3’X-tail would recruit the viral polymerase (NS5B) and other replication complex factors (both RNA and proteins)[92,109,111,113-117]. In this context, both the 5BSL3.2-IIId and 5BSL3.2-Alt interactions could be equally feasible. Swapping between them could contribute to the creation of a translational repressed state[25] and an enhanced replicative process[109]. The subsequent amplification of viral RNA molecules would displace the structural equilibrium between the 5BSL3.2-IIId and 5BSL3.2-Alt interactions toward the long-range IRES-CRE contact. This would increase the proportion of RNA genomes exposing the DLS motif in the apical loop of the dimerizable conformation, leading to the formation of dimeric genomic particles in the presence of the core chaperone protein[34].
Therefore, domain 5BSL3.2 would occupy the central position in a complex and dynamic interacting web that would help to bring the ends of the HCV genome into close proximity to support the formation of a biologically favoured close-loop topology. Swapping between different RNA structural partners through the viral cycle would thus control the course of the infectious process.
The search for novel conserved RNA structural units in viral genomes has been prompted in recent years by the appearance of bioinformatic tools that allow the study of the secondary structure of whole RNA genomes. Initial investigations based on the study of folding free energies in many positive stranded animal and plant viral RNA genomes identified extensive secondary structure regions that followed well-defined patterns[6,39]-the so-called GORS. They were initially related to different mechanisms for controlling viral replication, yet their prevalence appeared to be quite variable among different genera. For example, extensive base-pairing within the coding sequence was thermodynamically predicted for the hepacivirus genome, while in the closely related Pestivirus and Flavivirus genera this pattern was clearly absent. Since replication strategies are usually conserved among the members of a same family, it is unlikely that GORS work as fundamental base for the execution of the viral cycle. Remarkably, GORS are strongly associated to viral persistence[7], thus raising the question whether they can be involved in the suppression of innate intracellular defence mechanisms. Thermodynamic predictions and phylogenetic studies based on base-pairing rules have recently been combined with oligonucleotide probe accessibility and atomic force microscopy studies to investigate the link between theoretical predictions and the 3D conformation of viral genomes with and without GORS in solution[7]. The results showed that the HCV genome is a tightly compact molecule, in contrast to RNA genomes that lack GORS, such as that of poliovirus which folds pleomorphically, commonly involving long single stranded stretches. These studies have contributed to understand how RNA conformation could be related with a virus defence system. Thus, it seems likely that extensive folding areas could interfere with the antiviral cellular pathways triggered by double-stranded RNA, such as the interferon production during the initial infection, in an analogous manner to the expression of structured RNA transcripts by large DNA viruses[118]. Though much remains to be investigated about this phenomenon, it has an undeniable relevance for virus-host interactions.
During last years, the great advances in the fields of RNA structure determination by high-throughput techniques and bioinformatic tools have enabled the first pictures for the structural organization of the eukaryotic transcriptome, the so-called structure. In molecular virology, these advances have gained special relevance for their implication in the identification of functional RNA domains. These structurally conserved RNA elements interact with protein factors and other RNA domains to direct and regulate essential viral functions as well as switching between different steps of the viral cycle. Interfering with the functioning of these structural domains offers a potential means of treating viral infections, such as that caused by the HCV. Further implementations of the current methodologies will undoubtedly improve the identification and validation of functional RNA domains in the near future, thus extending our knowledge of RNA-mediated regulation not only in viral systems, but also in many cellular processes.
| 1. | Eigen M. Error catastrophe and antiviral strategy. Proc Natl Acad Sci USA. 2002;99:13374-13376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 237] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 2. | Wagner A, Stadler PF. Viral RNA and evolved mutational robustness. J Exp Zool. 1999;285:119-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 3. | Contreras AM, Hiasa Y, He W, Terella A, Schmidt EV, Chung RT. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J Virol. 2002;76:8505-8517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 144] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 4. | Churkin A, Cohen M, Shemer-Avni Y, Barash D. Bioinformatic analysis of the neutrality of RNA secondary structure elements across genotypes reveals evidence for direct evolution of genetic robustness in HCV. J Bioinform Comput Biol. 2010;8:1013-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Romero-López C, Berzal-Herranz A. Unmasking the information encoded as structural motifs of viral RNA genomes: a potential antiviral target. Rev Med Virol. 2013;23:340-354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 6. | Simmonds P, Tuplin A, Evans DJ. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: Implications for virus evolution and host persistence. RNA. 2004;10:1337-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 170] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 7. | Davis M, Sagan SM, Pezacki JP, Evans DJ, Simmonds P. Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian RNA viruses. J Virol. 2008;82:11824-11836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 81] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Zhu Y, Chen S. Antiviral treatment of hepatitis C virus infection and factors affecting efficacy. World J Gastroenterol. 2013;19:8963-8973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 37] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Simmonds P, McOmish F, Yap PL, Chan SW, Lin CK, Dusheiko G, Saeed AA, Holmes EC. Sequence variability in the 5’ non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J Gen Virol. 1993;74:661-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 284] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 10. | Thomson BJ, Finch RG. Hepatitis C virus infection. Clin Microbiol Infect. 2005;11:86-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 77] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Martell M, Esteban JI, Quer J, Genescà J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225-3229. [PubMed] |
| 12. | Farci P. Hepatitis C virus. The importance of viral heterogeneity. Clin Liver Dis. 2001;5:895-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4669] [Article Influence: 126.2] [Reference Citation Analysis (1)] |
| 14. | Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524-9528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 815] [Cited by in RCA: 827] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 15. | Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105-1113. [PubMed] |
| 16. | Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476-1483. [PubMed] |
| 17. | Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338-3344. [PubMed] |
| 18. | Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010-6020. [PubMed] |
| 19. | Wang TH, Rijnbrand RC, Lemon SM. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J Virol. 2000;74:11347-11358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Ito T, Tahara SM, Lai MM. The 3’-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789-8796. [PubMed] |
| 21. | McCaffrey AP, Ohashi K, Meuse L, Shen S, Lancaster AM, Lukavsky PJ, Sarnow P, Kay MA. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther. 2002;5:676-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3’-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293-1303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 74] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 23. | Song Y, Friebe P, Tzima E, Jünemann C, Bartenschlager R, Niepmann M. The hepatitis C virus RNA 3’-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol. 2006;80:11579-11588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 108] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 24. | Bung C, Bochkaeva Z, Terenin I, Zinovkin R, Shatsky IN, Niepmann M. Influence of the hepatitis C virus 3’-untranslated region on IRES-dependent and cap-dependent translation initiation. FEBS Lett. 2010;584:837-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Romero-López C, Berzal-Herranz A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell Mol Life Sci. 2012;69:103-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Ito T, Lai MM. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3’-untranslated sequence. Virology. 1999;254:288-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 27. | Fang JW, Moyer RW. The effects of the conserved extreme 3’ end sequence of hepatitis C virus (HCV) RNA on the in vitro stabilization and translation of the HCV RNA genome. J Hepatol. 2000;33:632-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Kong LK, Sarnow P. Cytoplasmic expression of mRNAs containing the internal ribosome entry site and 3’ noncoding region of hepatitis C virus: effects of the 3’ leader on mRNA translation and mRNA stability. J Virol. 2002;76:12457-12462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 29. | Romero-López C, Berzal-Herranz A. A long-range RNA-RNA interaction between the 5’ and 3’ ends of the HCV genome. RNA. 2009;15:1740-1752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 30. | Romero-López C, Barroso-Deljesus A, García-Sacristán A, Briones C, Berzal-Herranz A. The folding of the hepatitis C virus internal ribosome entry site depends on the 3’-end of the viral genome. Nucleic Acids Res. 2012;40:11697-11713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | Romero-López C, Barroso-Deljesus A, García-Sacristán A, Briones C, Berzal-Herranz A. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3’X-tail region. Nucleic Acids Res. 2014;42:567-582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 32. | Friebe P, Lohmann V, Krieger N, Bartenschlager R. Sequences in the 5’ nontranslated region of hepatitis C virus required for RNA replication. J Virol. 2001;75:12047-12057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 258] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 33. | Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3’ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol. 2002;76:5326-5338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 206] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 34. | Cristofari G, Ivanyi-Nagy R, Gabus C, Boulant S, Lavergne JP, Penin F, Darlix JL. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004;32:2623-2631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 95] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 35. | Ivanyi-Nagy R, Kanevsky I, Gabus C, Lavergne JP, Ficheux D, Penin F, Fossé P, Darlix JL. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res. 2006;34:2618-2633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 74] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 36. | Shetty S, Kim S, Shimakami T, Lemon SM, Mihailescu MR. Hepatitis C virus genomic RNA dimerization is mediated via a kissing complex intermediate. RNA. 2010;16:913-925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 37. | Sharma KK, de Rocquigny H, Darlix JL, Lavergne JP, Pénin F, Lessinger JM, Mély Y. Analysis of the RNA chaperoning activity of the hepatitis C virus core protein on the conserved 3’X region of the viral genome. Nucleic Acids Res. 2012;40:2540-2553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 38. | Smith DB, Simmonds P. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45:238-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 72] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 39. | Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA. 2002;8:824-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 104] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 40. | Kim YK, Lee SH, Kim CS, Seol SK, Jang SK. Long-range RNA-RNA interaction between the 5’ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA. 2003;9:599-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 41. | Beguiristain N, Robertson HD, Gómez J. RNase III cleavage demonstrates a long range RNA: RNA duplex element flanking the hepatitis C virus internal ribosome entry site. Nucleic Acids Res. 2005;33:5250-5261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 42. | McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci USA. 2007;104:2879-2884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 95] [Article Influence: 5.0] [Reference Citation Analysis (29)] |
| 43. | Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccalà G, Leroux-Roels G, Mavromara P, Bartenschlager R. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol. 2008;82:11503-11515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 84] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 44. | Lee H, Shin H, Wimmer E, Paul AV. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J Virol. 2004;78:10865-10877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 77] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 45. | You S, Stump DD, Branch AD, Rice CM. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol. 2004;78:1352-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 178] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 46. | Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 591] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 47. | Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 309] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 48. | Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 209] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 49. | Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959-1962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 386] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 50. | Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5’ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775-4782. [PubMed] |
| 51. | Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795-805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 125] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 52. | Brown EA, Zhang H, Ping LH, Lemon SM. Secondary structure of the 5’ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041-5045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 302] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 53. | Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 182] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 54. | Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955-968. [PubMed] |
| 55. | Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456-1466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 90] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 56. | Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 5’ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165-1174. [PubMed] |
| 57. | Pestova TV, Hellen CU. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology. 1999;258:249-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 58. | Lyons AJ, Lytle JR, Gomez J, Robertson HD. Hepatitis C virus internal ribosome entry site RNA contains a tertiary structural element in a functional domain of stem-loop II. Nucleic Acids Res. 2001;29:2535-2541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 59. | Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 225] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 60. | Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol. 2000;74:6242-6250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 130] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 61. | Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8:1045-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 62. | Kalliampakou KI, Psaridi-Linardaki L, Mavromara P. Mutational analysis of the apical region of domain II of the HCV IRES. FEBS Lett. 2002;511:79-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 63. | Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA. 2004;101:16990-16995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 142] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 64. | Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 204] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 65. | Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 267] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 66. | Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit’s decoding groove. RNA. 2011;17:1258-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 67. | Lafuente E, Ramos R, Martínez-Salas E. Long-range RNA-RNA interactions between distant regions of the hepatitis C virus internal ribosome entry site element. J Gen Virol. 2002;83:1113-1121. [PubMed] |
| 68. | Malygin AA, Kossinova OA, Shatsky IN, Karpova GG. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013;41:8706-8714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 69. | Buratti E, Tisminetzky S, Zotti M, Baralle FE. Functional analysis of the interaction between HCV 5’UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 1998;26:3179-3187. [PubMed] |
| 70. | Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 71. | Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JY, Baroudy BM. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J Virol. 2000;74:10430-10437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 93] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 72. | Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009;37:1141-1151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 73. | Klinck R, Westhof E, Walker S, Afshar M, Collier A, Aboul-Ela F. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA. 2000;6:1423-1431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 74. | Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7:1105-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 167] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 75. | Berry KE, Waghray S, Doudna JA. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559-1569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 76. | Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 154] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 77. | Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3’ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74:2046-2051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 506] [Cited by in RCA: 489] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 78. | Yi M, Lemon SM. 3’ nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol. 2003;77:3557-3568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 150] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 79. | Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3’ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363-3371. [PubMed] |
| 80. | You S, Rice CM. 3’ RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J Virol. 2008;82:184-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 101] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 81. | Gontarek RR, Gutshall LL, Herold KM, Tsai J, Sathe GM, Mao J, Prescott C, Del Vecchio AM. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3’NTR of the HCV RNA genome. Nucleic Acids Res. 1999;27:1457-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 82. | Chang KS, Luo G. The polypyrimidine tract-binding protein (PTB) is required for efficient replication of hepatitis C virus (HCV) RNA. Virus Res. 2006;115:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 83. | Spångberg K, Goobar-Larsson L, Wahren-Herlenius M, Schwartz S. The La protein from human liver cells interacts specifically with the U-rich region in the hepatitis C virus 3’ untranslated region. J Hum Virol. 1999;2:296-307. [PubMed] |
| 84. | Petrik J, Parker H, Alexander GJ. Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly(U) tract of the 3’ non-coding region of hepatitis C virus genomic RNA. J Gen Virol. 1999;80:3109-3113. [PubMed] |
| 85. | Luo G. Cellular proteins bind to the poly(U) tract of the 3’ untranslated region of hepatitis C virus RNA genome. Virology. 1999;256:105-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 86. | Wang H, Shen XT, Ye R, Lan SY, Xiang L, Yuan ZH. Roles of the polypyrimidine tract and 3’ noncoding region of hepatitis C virus RNA in the internal ribosome entry site-mediated translation. Arch Virol. 2005;150:1085-1099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 87. | Kim M, Kim H, Cho SP, Min MK. Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA. J Virol. 2002;76:6944-6956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 88. | Shim JH, Larson G, Wu JZ, Hong Z. Selection of 3’-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J Virol. 2002;76:7030-7039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 89. | Murayama A, Weng L, Date T, Akazawa D, Tian X, Suzuki T, Kato T, Tanaka Y, Mizokami M, Wakita T. RNA polymerase activity and specific RNA structure are required for efficient HCV replication in cultured cells. PLoS Pathog. 2010;6:e1000885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 90. | Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem. 2005;280:36417-36428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 205] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 91. | Harris D, Zhang Z, Chaubey B, Pandey VN. Identification of cellular factors associated with the 3’-nontranslated region of the hepatitis C virus genome. Mol Cell Proteomics. 2006;5:1006-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 79] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 92. | Tingting P, Caiyun F, Zhigang Y, Pengyuan Y, Zhenghong Y. Subproteomic analysis of the cellular proteins associated with the 3’ untranslated region of the hepatitis C virus genome in human liver cells. Biochem Biophys Res Commun. 2006;347:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 93. | Kim CS, Seol SK, Song OK, Park JH, Jang SK. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J Virol. 2007;81:3852-3865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 94. | Scheller N, Mina LB, Galão RP, Chari A, Giménez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Díez J. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci USA. 2009;106:13517-13522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 116] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 95. | Weinlich S, Hüttelmaier S, Schierhorn A, Behrens SE, Ostareck-Lederer A, Ostareck DH. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3‘UTR. RNA. 2009;15:1528-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 96. | Yu KL, Jang SI, You JC. Identification of in vivo interaction between Hepatitis C Virus core protein and 5’ and 3’ UTR RNA. Virus Res. 2009;145:285-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 97. | Bai Y, Zhou K, Doudna JA. Hepatitis C virus 3’UTR regulates viral translation through direct interactions with the host translation machinery. Nucleic Acids Res. 2013;41:7861-7874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 98. | Friebe P, Boudet J, Simorre JP, Bartenschlager R. Kissing-loop interaction in the 3’ end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79:380-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 296] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 99. | Tuplin A, Struthers M, Simmonds P, Evans DJ. A twist in the tail: SHAPE mapping of long-range interactions and structural rearrangements of RNA elements involved in HCV replication. Nucleic Acids Res. 2012;40:6908-6921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 100. | Ina Y, Mizokami M, Ohba K, Gojobori T. Reduction of synonymous substitutions in the core protein gene of hepatitis C virus. J Mol Evol. 1994;38:50-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 101. | Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 2001;20:3840-3848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 211] [Cited by in RCA: 206] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 102. | Varaklioti A, Vassilaki N, Georgopoulou U, Mavromara P. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J Biol Chem. 2002;277:17713-17721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 113] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 103. | Vassilaki N, Kalliampakou KI, Mavromara P. Differences in the expression of the hepatitis C virus core+1 open reading frame between a nuclear and a cytoplasmic expression system. J Gen Virol. 2008;89:222-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 104. | Díaz-Toledano R, Ariza-Mateos A, Birk A, Martínez-García B, Gómez J. In vitro characterization of a miR-122-sensitive double-helical switch element in the 5’ region of hepatitis C virus RNA. Nucleic Acids Res. 2009;37:5498-5510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 105. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1993] [Cited by in RCA: 1998] [Article Influence: 95.1] [Reference Citation Analysis (0)] |
| 106. | Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300-3310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 492] [Cited by in RCA: 531] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 107. | Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716-7729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 108. | Hofacker IL, Fekete M, Flamm C, Huynen MA, Rauscher S, Stolorz PE, Stadler PF. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 1998;26:3825-3836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 91] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 109. | Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J Virol. 2008;82:9008-9022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 83] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 110. | Zhang J, Yamada O, Sakamoto T, Yoshida H, Araki H, Murata T, Shimotohno K. Inhibition of hepatitis C virus replication by pol III-directed overexpression of RNA decoys corresponding to stem-loop structures in the NS5B coding region. Virology. 2005;342:276-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 111. | Ito T, Lai MM. Determination of the secondary structure of and cellular protein binding to the 3’-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698-8706. [PubMed] |
| 112. | Shetty S, Stefanovic S, Mihailescu MR. Hepatitis C virus RNA: molecular switches mediated by long-range RNA-RNA interactions? Nucleic Acids Res. 2013;41:2526-2540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 113. | Banerjee R, Dasgupta A. Specific interaction of hepatitis C virus protease/helicase NS3 with the 3’-terminal sequences of viral positive- and negative-strand RNA. J Virol. 2001;75:1708-1721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 114. | Yi M, Lemon SM. Structure-function analysis of the 3’ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA. 2003;9:331-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 115. | Foster TL, Belyaeva T, Stonehouse NJ, Pearson AR, Harris M. All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J Virol. 2010;84:9267-9277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 99] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 116. | Kanamori H, Yuhashi K, Ohnishi S, Koike K, Kodama T. RNA-dependent RNA polymerase of hepatitis C virus binds to its coding region RNA stem-loop structure, 5BSL3.2, and its negative strand. J Gen Virol. 2010;91:1207-1212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 117. | Oakland TE, Haselton KJ, Randall G. EWSR1 binds the hepatitis C virus cis-acting replication element and is required for efficient viral replication. J Virol. 2013;87:6625-6634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 118. | Mathews MB. Structure, function, and evolution of adenovirus virus-associated RNAs. Curr Top Microbiol Immunol. 1995;199:173-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
P- Reviewers: Bock T, Lee sw S- Editor: Ma YJ L- Editor: A E- Editor: Liu SQ