Revised: May 14, 2013
Accepted: May 20, 2013
Published online: May 27, 2013
Processing time: 118 Days and 6.4 Hours
Microduplications are normally invisible under microscopy and were not recognized before chromosomal microarray testing was available. Although it is difficult to confirm the orientation of duplicated segments by standard fluorescence in situ hybridization (FISH), our data indicates that fiber-FISH analysis has the potential to reveal the orientation of duplicated and triplicated segments of chromosomes. Recurrent microduplications reciprocal to microdeletions show tandem orientations of the duplicated segments, which is consistent with a non-allelic homologous recombination mechanism. Several random duplications showed tandem configurations and inverted duplications are rare. Further analysis is required to fully elucidate the basic mechanisms underlying such duplications/triplications.
Core tip: Fiber-fluorescence in situ hybridization analysis has the potential to reveal the orientation of duplicated and triplicated segments of chromosomes. Previously, we reported that interstitial duplications were aligned in tandem configurations, supporting the hypothesized mechanism of non-allelic homologous recombination; however, there were rare cases of inverted duplications. Further analysis is therefore required to fully elucidate the basic mechanisms underlying such duplications/triplications.
-
Citation: Yamamoto T, Shimada S, Shimojima K. Fiber-fluorescence
in situ hybridization analyses as a diagnostic application for orientation of microduplications. World J Med Genet 2013; 3(2): 5-8 - URL: https://www.wjgnet.com/2220-3184/full/v3/i2/5.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v3.i2.5
Chromosomal microduplication is a type of chromosomal anomaly. Although several chromosomal duplications have been reported previously, small microduplications that were invisible under the microscope were not recognized before chromosomal microarray testing was available. The only known microduplications were those involving specific genes, such as the peripheral myelin protein 22 gene (PMP22) and the proteolipid protein 1 gene (PLP1). Previously, we knew that patients with PMP22 mutations manifest Charcot-Marie-Tooth disease [Mendelian Inheritance in Man (MIM) #118220] and patients with PLP1 mutations show Pelizaeus-Merzbacher disease (PMD; MIM #312080); microduplications of these genes were revealed under the hypothesis that copy number gain of these genes may be related to disease occurrence[1]. Before the availability of chromosomal microarray testing, a targeting system was used to detect such duplications, including fluorescence in situ hybridization (FISH), quantitative polymerase chain reaction (PCR) and multiple ligation probe amplification (MLPA) (Table 1).
| Characters | Advantages | Disadvantages |
| Fluorescence in situ hybridization | Numbers of the signals correspond to the genomic copy numbers | Small duplications cannot be detected |
| Quantitative polymerase chain reaction | Wide dynamic range | Primer designs are required for each targets |
| Many samples can be analyzed at once | ||
| Multiple ligation probe amplification | Multiple targets can be analyzed at once | Original primer designs are required for specific regions |
| Chromosomal microarray testing | Accurate and high resolution | Chromosomal structures including balanced translocations cannot be analyzed |
| Comprehensive |
It is possible to detect the numbers of the targeted signals by FISH - whole subtelomeres can be analyzed in this manner[2]. Specific regions have already been analyzed previously using this method; e.g., the 22q11.2 region for DiGeorge syndrome and the 7q11.2 region for Williams syndrome. Although FISH is a powerful tool for detecting deletions of targeted regions, the biggest disadvantage is that it is difficult to detect small duplications owing to overlapping signals.
Generally, PCR cannot be used to quantify amplified fragments owing to saturation of amplification. However, in cases of linear amplification, quantity can be estimated by using a real-time PCR monitoring system. The chief feature of this system is that the target is fixed at the start of analysis[3]. Although MLPA is based on a PCR system, the targets of amplification are fused probes included in the buffer. Because various sizes of the amplicons can be included in the same tube, multiplexed targets can be measured simultaneously[4] and whole subtelomeric regions can be analyzed comprehensively[5].
At present, it is easier to detect genomic copy number aberrations by using chromosomal microarray testing as a comprehensive method. We subsequently realized that there are numerous microchromosomal duplications that are genetic causes for various diseases.
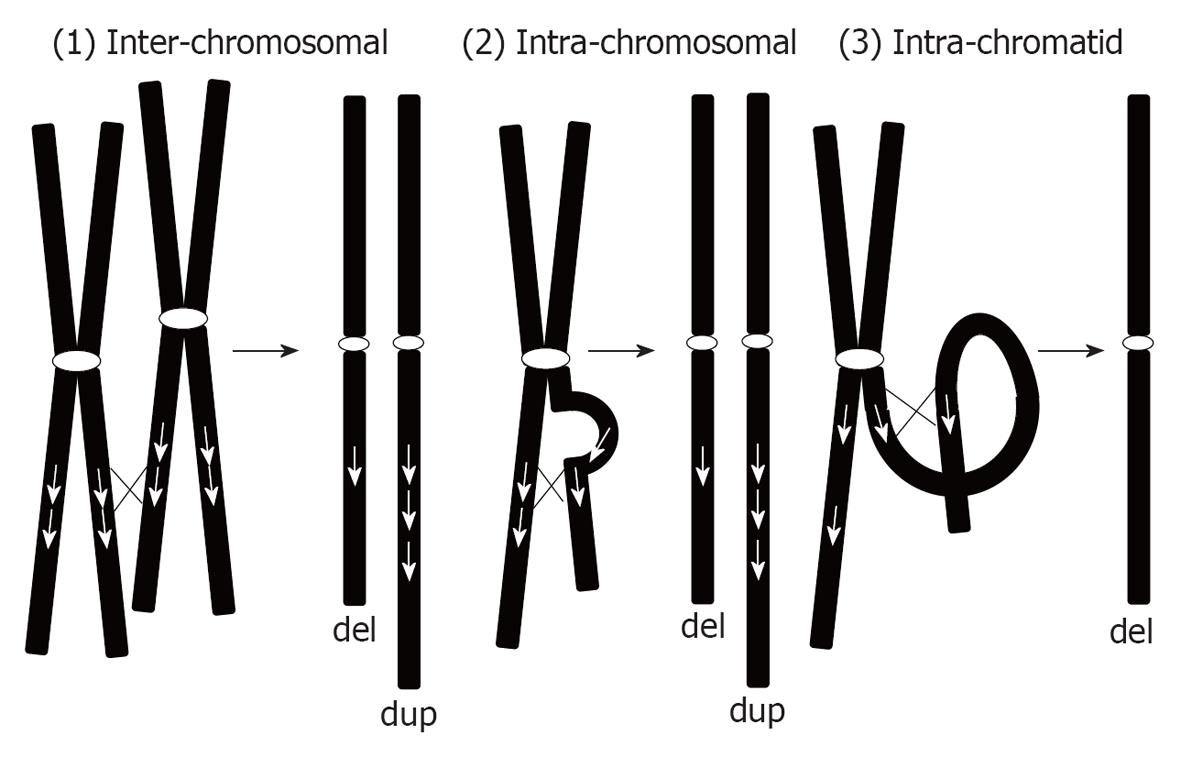
We can classify duplications into 2 types: recurrent microduplications and random microduplications (Table 2). Recurrent microduplications are caused by non-allelic homologous recombination (NAHR) mediated by low-copy repeats. Consequently, such microduplications are reciprocal to the microdeletions caused by the NAHR mechanism. According to this mechanism, chromosomal aberrations can occur in 3 ways[6]: (1) inter-chromosomal; (2) intra-chromosomal; and (3) intra-chromatid (Figure 1). As shown in Figure 1, inter-chromosomal and intra-chromosomal exchanges can create both deletions and duplications equally. However, intra-chromatid exchange only creates microdeletion and not microduplication. Thus, microduplications created by NAHR are definitely a consequence of inter-chromosomal or intra-chromosomal exchange. Both processes create duplications in tandem orientation. However, few studies have confirmed the hypothesized tandem configurations of the duplication.
| Characteristic | Recurrent microduplications | Random microduplications |
| Mechanism | Non-allelic homologous recombination | Random |
| Mediated by locus control regions | ||
| Characteristics | Reciprocal to common deletions | Random |
| Uniformed size |
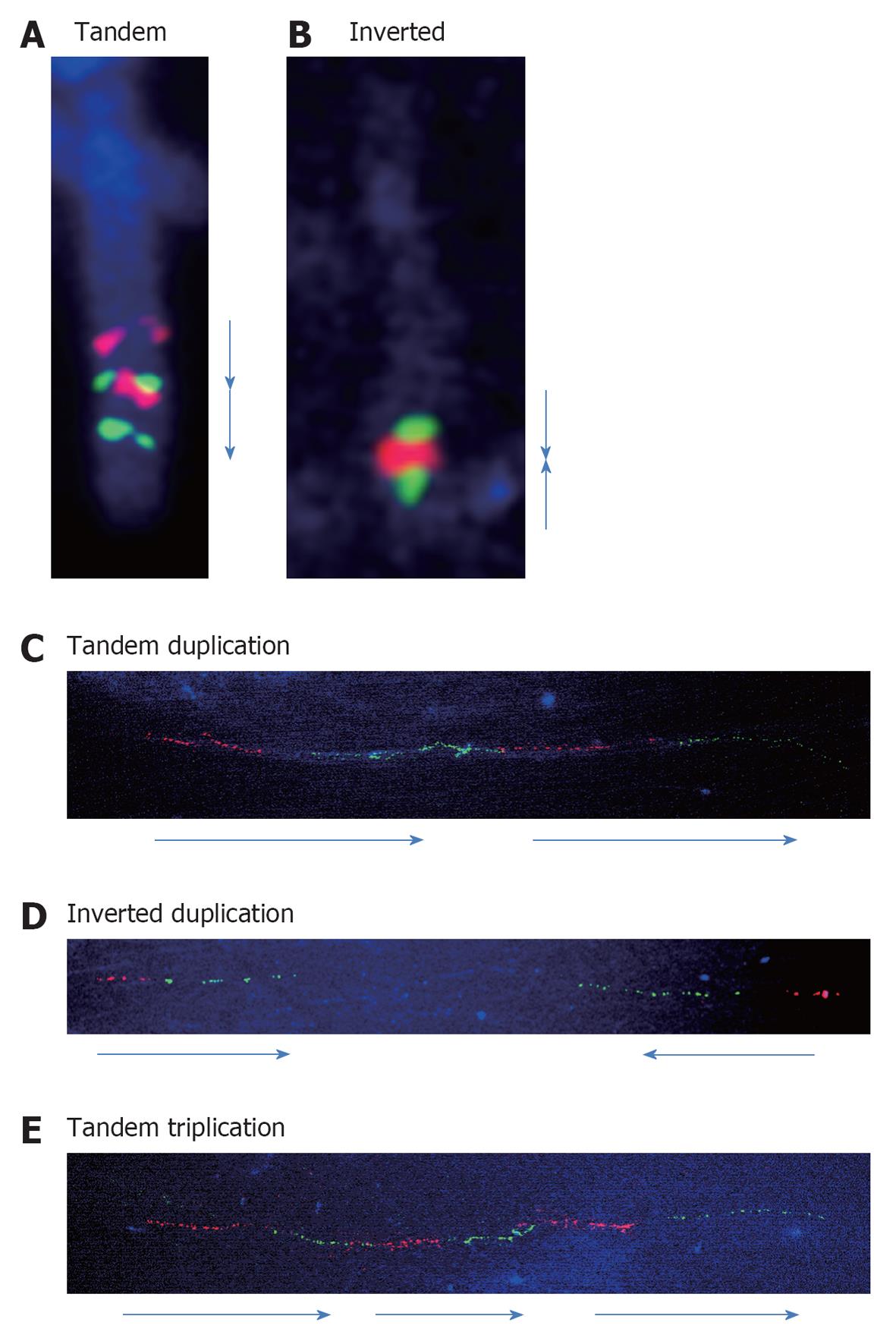
Previously, many efforts have been made to detect orientations of the duplication by FISH. The combined use of 2 different colored probes can detect the orientations of the chromosomal fragments. We analyzed the orientations of the duplicated segments and many showed tandem configurations. As shown in Figure 2A, an interstitial duplication of 2q32.1-q33.3 was inserted in a contiguous tandem configuration[7].
Compared to such interstitial duplications, subtelomere duplications are different because they are most commonly a result of U-type exchange between the sister chromatids during the meiotic process[8]. We observed inverted duplication and deletion in the 10q26 region[9]. Inverted orientation was confirmed by FISH in this case (Figure 2B).
As mentioned above, in the cases of large duplications it is possible to determine the orientations of the duplicated segments; however, it is difficult to determine orientation for small duplications owing to overlapping signals in metaphase spreads. Consequently, interphase nuclei were used as an alternative to determine the alignments of the signals. Lee et al[10] analyzed the orientations of the duplications involving PLP1 in patients with PMD using interphase nuclei. They confirmed that many duplications of this region were aligned in tandem orientations. However, there is the limitation that directions cannot be detected accurately. To compensate for this, several nuclei from the same patients have to be checked.
Chromosomes are composed of 3-dimentional structures that consist of DNA and histones. Thus, targeted signals can be overlapped. In such cases, fiber-FISH analysis has the advantage of being able to detect the orientations of the duplicated segments accurately[11-13] (“fiber” means DNA fibers).
DNA fiber specimens can be prepared after separating chromatin structures by surfactants. To perform fiber-FISH analysis, traditional Carnoy fixation can be used. The method is as follows: Carnoy-fixed samples should be mounted on the surface of the glass slides and the slides should be immediately dipped into sodium dodecyl sulfate solution and then slowly pulled out. Consequently, the separated DNA fibers can be fixed onto the surface of the glass slides.
The same probes used for standard FISH can be used for fiber-FISH. Compared to standard FISH, targeted signal intensity is extremely weak. Because live signals are not visible by microscopy, long exposure time is required for signal capture. To capture standard FISH pictures, interphase nucleus or metaphase labeled by DAPI can be used as landmarks; however, there are no interphase nucleus and metaphase in fiber-FISH specimens. Thus, it is necessary to search the entire surface of the slide to detect signals.
Not all fibers show the same rate of extension on the same slide. This is dependent on the duration of the dip into the surfactants solution. If the length of the targeted fiber was too long to be captured in a single field of view, not all the target signals can be captured in the same field of view. It also depends on the size of the targeted duplications. If signals can be successfully captured, the directions of the fragments can be determined.
In terms of the clinical point of view, detection of microduplication orientation is not a suitable strategy for clinical analyses. Thus, fiber-FISH analysis is a specialized method for research work.
Previously, we analyzed the directions of the duplicated segments of the 22q11.2 region. The 4 duplications of the 22q11.2 region, identified by chromosomal microarray testing, showed tandem configurations of the duplicated segments, supporting the hypothesized mechanism of NAHR[11]. We analyzed 7 and 4 samples with the duplications in the region of PLP1 and the methyl CpG binding protein 2 gene, respectively[12-14], and all of the duplications showed tandem configurations as seen in the 22q11.2 region (Figure 2C), although the duplications in the region of PLP1 are considered to be created by fork stalling and template switching and not by NAHR[15].
Almost all duplications analyzed previously showed tandem configurations, as seen in cases of large duplications visible by general FISH analysis. The only exception was a benign copy number gain identified at the Xp22.31 region that included the steroid sulfatase gene (Figure 2D)[12]. This may indicate that the mechanism of the copy number gain in the benign region may be different from that of the pathological CNV.
Triplications are rare chromosomal aberrations which can be classified into 2 types: (1) triplication embedded into duplicated segments; and (2) triplications not embedded into duplicated segments[16]. Because sufficient data are not available owing to the scarcity of these triplications, the mechanisms underlying them have seldom been analyzed. The clearest evidence is that some of the triplications embedded into duplicated segments are caused by the duplication-inverted triplication-duplication mechanism revealed by Carvalho et al[17] and Shimojima et al[18]. Compared to this, a tandem triplication was confirmed in another case involving the platelet-activating factor acetylhydrolase 1b regulatory subunit gene (Figure 2E)[19].
In conclusion, as a result of the wider adoption of chromosomal microarray testing as a diagnostic tool, many genomic copy number gains were found to cause multiple congenital anomalies and intellectual impairments. Thus, this is just a starting point to understanding the mechanism of such genomic copy number gains. For this purpose, we should accumulate more cases of duplications and triplications and analyze the orientations of the segments.
| 1. | Yamamoto T, Shimojima K. Pelizaeus-Merzbacher disease as a chromosomal disorder. Congenit Anom (Kyoto). 2013;53:3-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 2. | Ravnan JB, Tepperberg JH, Papenhausen P, Lamb AN, Hedrick J, Eash D, Ledbetter DH, Martin CL. Subtelomere FISH analysis of 11 688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J Med Genet. 2006;43:478-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 275] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 3. | Ceulemans S, van der Ven K, Del-Favero J. Targeted screening and validation of copy number variations. Methods Mol Biol. 2012;838:311-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Warshawsky I, Chernova OB, Hübner CA, Stindl R, Henneke M, Gal A, Natowicz MR. Multiplex ligation-dependent probe amplification for rapid detection of proteolipid protein 1 gene duplications and deletions in affected males and carrier females with Pelizaeus-Merzbacher disease. Clin Chem. 2006;52:1267-1275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Pohovski LM, Dumic KK, Odak L, Barisic I. Multiplex ligation-dependent probe amplification workflow for the detection of submicroscopic chromosomal abnormalities in patients with developmental delay/intellectual disability. Mol Cytogenet. 2013;6:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 426] [Cited by in RCA: 461] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 7. | Usui D, Shimada S, Shimojima K, Sugawara M, Kawasaki H, Shigematu H, Takahashi Y, Inoue Y, Imai K, Yamamoto T. Interstitial duplication of 2q32.1-q33.3 in a patient with epilepsy, developmental delay, and autistic behavior. Am J Med Genet A. 2013;161A:1078-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Rowe LR, Lee JY, Rector L, Kaminsky EB, Brothman AR, Martin CL, South ST. U-type exchange is the most frequent mechanism for inverted duplication with terminal deletion rearrangements. J Med Genet. 2009;46:694-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 9. | Kibe T, Mori Y, Okanishi T, Shimojima K, Yokochi K, Yamamoto T. Two concurrent chromosomal aberrations involving interstitial deletion in 1q24.2q25.2 and inverted duplication and deletion in 10q26 in a patient with stroke associated with antithrombin deficiency and a patent foramen ovale. Am J Med Genet A. 2011;155A:215-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Lee JA, Inoue K, Cheung SW, Shaw CA, Stankiewicz P, Lupski JR. Role of genomic architecture in PLP1 duplication causing Pelizaeus-Merzbacher disease. Hum Mol Genet. 2006;15:2250-2265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | Shimojima K, Okamoto N, Inazu T, Yamamoto T. Tandem configurations of variably duplicated segments of 22q11.2 confirmed by fiber-FISH analysis. J Hum Genet. 2011;56:810-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | Shimojima K, Inoue T, Hoshino A, Kakiuchi S, Watanabe Y, Sasaki M, Nishimura A, Takeshita-Yanagisawa A, Tajima G, Ozawa H. Comprehensive genetic analyses of PLP1 in patients with Pelizaeus-Merzbacher disease applied by array-CGH and fiber-FISH analyses identified new mutations and variable sizes of duplications. Brain Dev. 2010;32:171-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 13. | Shimada S, Okamoto N, Ito M, Arai Y, Momosaki K, Togawa M, Maegaki Y, Sugawara M, Shimojima K, Osawa M. MECP2 duplication syndrome in both genders. Brain Dev. 2013;35:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Shimojima K, Inoue T, Imai Y, Arai Y, Komoike Y, Sugawara M, Fujita T, Ideguchi H, Yasumoto S, Kanno H. Reduced PLP1 expression in induced pluripotent stem cells derived from a Pelizaeus-Merzbacher disease patient with a partial PLP1 duplication. J Hum Genet. 2012;57:580-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 15. | Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849-853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 364] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 16. | Yamamoto T, Matsuo M, Shimada S, Sangu N, Shimojima K, Aso S, Saito K. De novo triplication of 11q12.3 in a patient with developmental delay and distinctive facial features. Mol Cytogenet. 2013;6:15. [PubMed] |
| 17. | Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43:1074-1081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 18. | Shimojima K, Mano T, Kashiwagi M, Tanabe T, Sugawara M, Okamoto N, Arai H, Yamamoto T. Pelizaeus-Merzbacher disease caused by a duplication-inverted triplication-duplication in chromosomal segments including the PLP1 region. Eur J Med Genet. 2012;55:400-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Shimojima K, Sugiura C, Takahashi H, Ikegami M, Takahashi Y, Ohno K, Matsuo M, Saito K, Yamamoto T. Genomic copy number variations at 17p13.3 and epileptogenesis. Epilepsy Res. 2010;89:303-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
P- Reviewers Corrales FJ, Li CJ, Petmitr S, Sipos F, Sterlacci W, Tanabe S S- Editor Gou SX L- Editor Roemmele A E- Editor Zheng XM