Published online Sep 20, 2024. doi: 10.5496/wjmg.v12.i1.93011
Revised: May 23, 2024
Accepted: July 2, 2024
Published online: September 20, 2024
Processing time: 216 Days and 13.4 Hours
Circular RNAs (circRNAs), a new star of noncoding RNAs, are a group of endogenous RNAs that form a covalently closed circle and occur widely in the mammalian genome. Most circRNAs are conserved throughout species and fre
Core Tip: This opinion review covers the biogenesis and metabolism of circular RNAs (circRNAs) as well as their functions and potential roles in major human diseases. We review major peer-reviewed articles published in the field of cirRNAs and the involvement of this class of noncoding RNAs in major human diseases. The role of circRNAs as molecular markers or potential targets will provide promising application perspectives, such as in early diagnoses, better treatment plans, the
- Citation: Sharma A, Bansal C, Sharma KL, Kumar A. Circular RNA: The evolving potential in the disease world. World J Med Genet 2024; 12(1): 93011
- URL: https://www.wjgnet.com/2220-3184/full/v12/i1/93011.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v12.i1.93011
The human genome is defined as “blueprint” for human life and is composed of DNA. The intermediate molecules arising from the genome, known as RNA, help manufacture crucial biomolecules such as proteins, which perform cellular processes. After sequencing the human genome, researchers surprisingly found that approximately 95% of the genome does not code for proteins. Since this is noncoding DNA, it originally appeared to have an unknown biological function, and many scientists referred to it as junk DNA or garbage of the human genome. However, there are multiple molecular milestones after the discovery of the function of the noncoding DNA, especially the regions of genome, which eventually get transcribed to RNA. Hence, these RNAs have been randomly designated as “intergenic RNA”, “long non-coding RNAs” etc., which initially led to their underrepresentation in molecular biology. Circular RNAs (circRNAs) are one of the great surprise discoveries that include a huge group of non-coding RNAs that are produced by an unconventional splicing method, also known as a non-canonical splicing event. Non-canonical splicing is called backsplicing, through which a downstream splice-donor site is covalently linked to an upstream splice-acceptor site. Previously mysterious circRNAs have been noted to impact gene expression by acting as microRNA (miRNA) sponges. Some circRNA mo
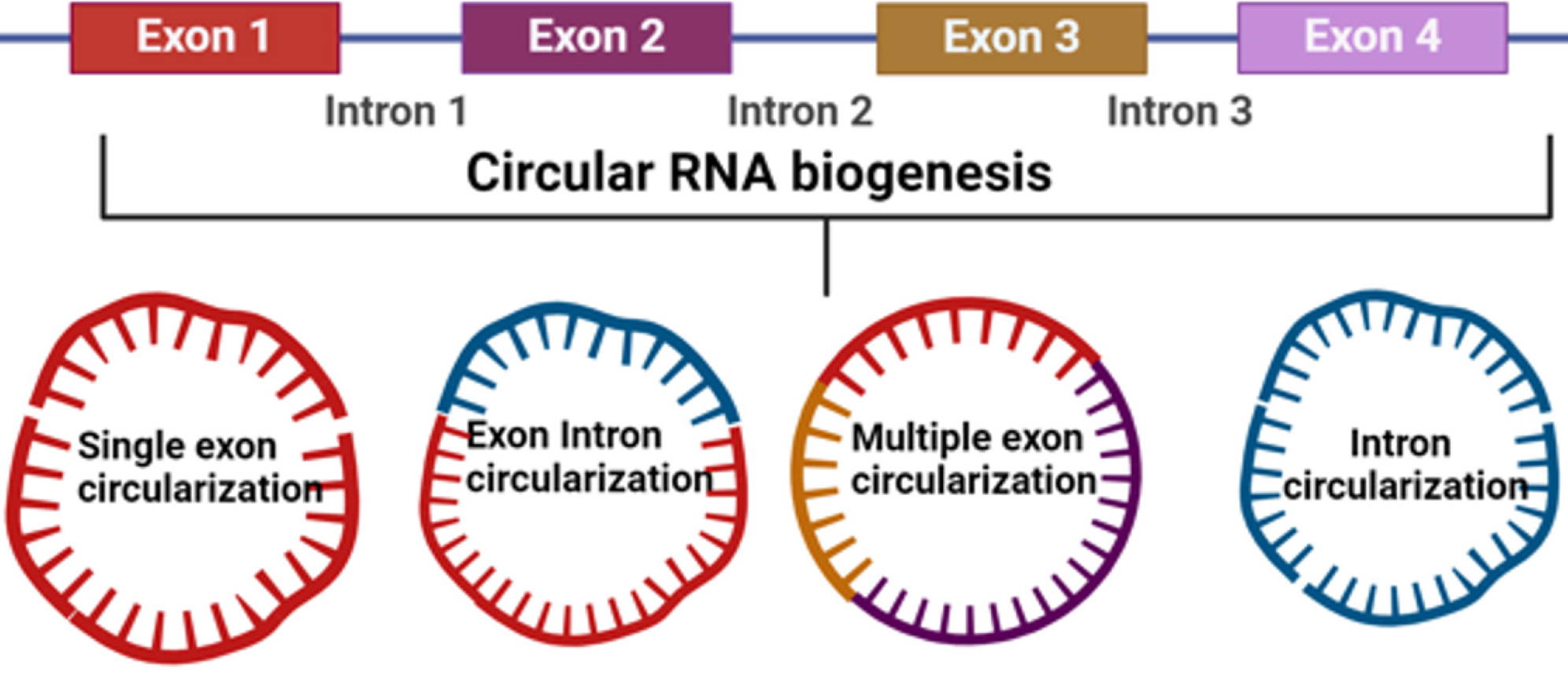
The noncoding RNAs (ncRNAs) can be characterized into two subdivisions, the housekeeper ncRNAs (rRNA, tRNA, snRNA, and snoRNA) and regulatory ncRNAs[2-4]. The regulatory ncRNAs are classified by length of transcript which comprises small ncRNAs < 200 bp (siRNA and miRNAs) and lnRNAs (transcript > 200 bp). CircRNAs are part of lnRNAs that emerged as a new class of endogenous RNAs and exist extensively in mammalian cells, which are emerging as crucial elements of cellular homeostasis. Figure 1 shows the circRNA biogenesis through various mechanisms.
CircRNAs are different from linear RNAs as the 3' and 5' ends typically present in an RNA molecule are merged. This characteristic feature has laid the foundation of various newly discovered functions of circRNAs. Many circRNAs evolve from protein-coding genes which do not code for any proteins themselves. Instead, they act as potential regulators of gene expression by various known and unknown biological processes. The first human circRNA was discovered around 32 years ago by Nigro et al[5] in 1991 from spliced transcripts of a candidate tumor suppressor gene, DCC[5]. This novel discovered RNA product had been considered to be a result of splicing errors during transcription[6]. Since advances in high throughput sequencing techniques, many circRNAs have been discovered, though there are still a lot to be dis
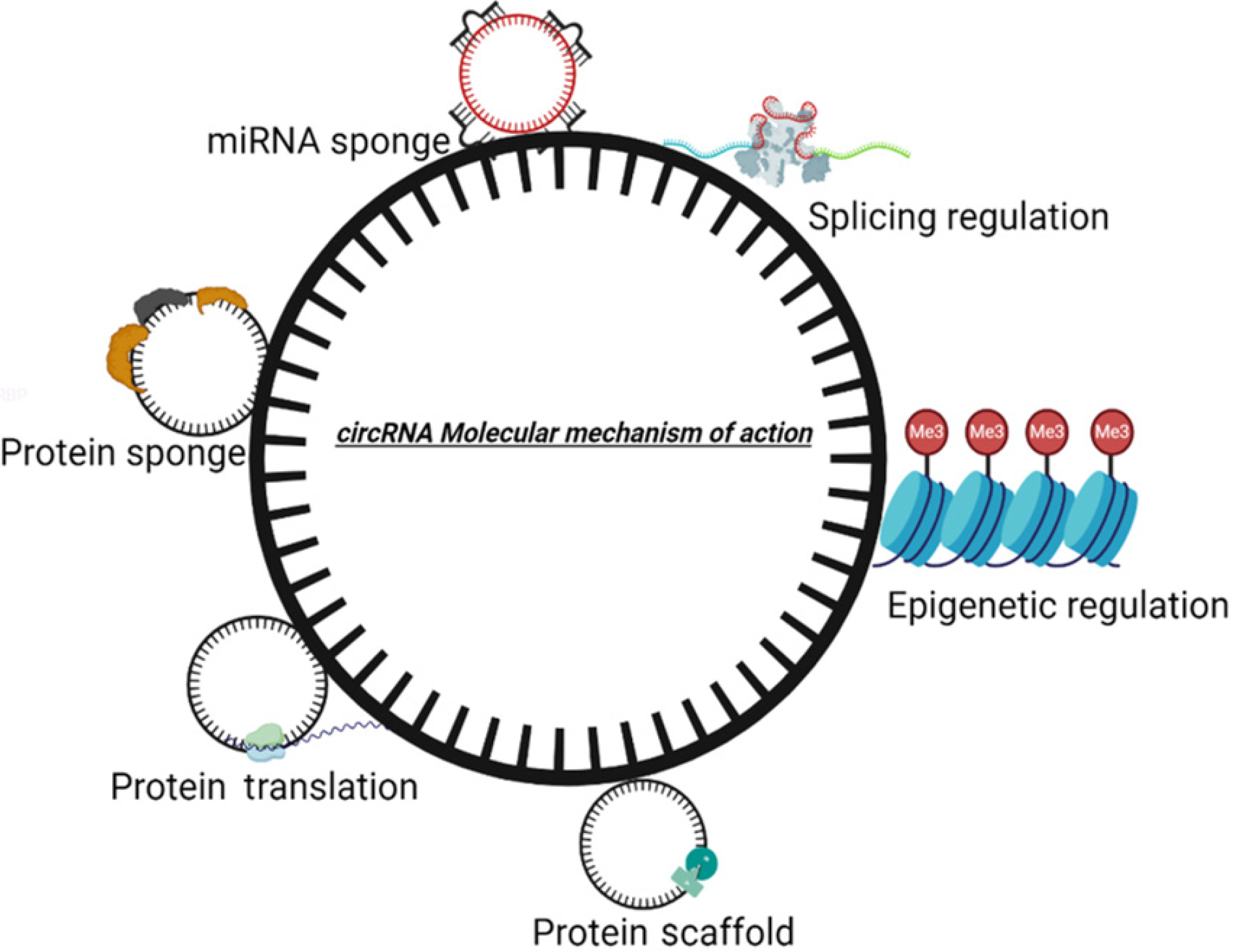
CircRNAs have remarkable potential in regulating the function of a gene. The various mechanism by which circRNAs are involved in controlling genetic events are sponging miRNA, interacting with RNA-binding protein (RBP), regulating transcription, regulating splicing, getting translated into proteins, and epigenetic regulation (Figure 2).
The major role of circRNAs as strong posttranscriptional regulators of gene expression is via acting as miRNAs sponges. They might also possibly sponge RBPs. CircRNAs throw genetics for a loop as they not only can act as sponges for miRNAs originating outside the cell but also have various other surprising functions including possible binding sites for viral miRNAs[10]. The combined analysis of high throughput transcriptome data coupled with deeper bioinformatic analyses characterizes a powerful approach to illuminate likely biological functions of ribonucleoprotein complexes. By examining circRNAs predominantly expressed in the brains of humans and mice, researchers at Aarhus University in Denmark, found that some circRNA molecules were blocking miR-7 (a type of miRNA) that usually inhibits expression of certain mRNAs. Therefore, the circRNA was controlling the activity of the blocker, increasing the expression of miR-7’s target genes[11]. A research article studied the expression profile of circRNAs using advanced microarray technology and demonstrated the changing expression profiles of 24 circRNAs and 37 miRNAs often altered at each stage of osteoclast differentiation during osteoclastogenesis[12]. This shows a great involvement of circRNAs in developmental biology. Nevertheless, the universal properties of circRNAs are not recognized yet. Because circRNAs do not have 5' or 3' end, they are resistant to exonuclease-mediated degradation and are presumably more stable than most linear RNAs in cells[13].
CircRNAs are mainly known to function as molecular sponges or decoys for miRNAs and controlling gene expression. CircRNAs can impact protein function either by sequestering the proteins and hence modifying their functional potential or via acting as a scaffold for protein-protein interaction. CircRNAs can interact with other RNA molecules and act as regulators of gene expression by interacting with RNA-binding proteins and modulating their functions. CircRNAs are also translated into proteins known as circRNA-derived proteins. Moreover, circRNAs have shown potential as diag
| Body system | Disease | circRNA | Expression | Ref. |
| Cardiovascular | Myocardial infarction | circRNA_081881 | Down | [43] |
| MICRA | Up | |||
| Cardiac fibrosis | circRNA_010567 | Up | [44] | |
| Congenital heart disease | hsa_circ_004183 | Down | [45] | |
| hsa_circ_079265 | ||||
| hsa_circ_105039 | ||||
| Hypertension | hsa_circ_0037911 | Up | [46] | |
| Cardiomyopathy | circDNAJC6 | Down | [47] | |
| circTMEM56 | ||||
| circMBOAT2 | ||||
| Heart failure | hsa_circ_0062960 | Up | [48] | |
| Coronary artery disease | hsa_circ_0124644 | Down | [49] | |
| hsa_circ_0001879 | Down | [50] | ||
| hsa_circ_0004104 | Down | [51] | ||
| hsa_circ_0001445 | Up | |||
| Atrial fibrillation | hsa_circ_025016 | Up | [52] | |
| Central nervous system | Moyamoya disease | hsa_circRNA_089761 | Down | [53] |
| hsa_circRNA_100914 | ||||
| hsa_circRNA_089763 | ||||
| Temporal lobe epilepsy | circ-EFCAB2 | Up | [54] | |
| circ-DROSHA | Down | [55] | ||
| circRNA-0067835 | Down | |||
| Alzheimer’s disease | CDR1as/ciRS-7 | Down | [56,57] | |
| circPVT1 | Up | [58] | ||
| Parkinson’s disease | CDR1as/ciRS-7 | Up | [59] | |
| circzip-2 | ||||
| Multiple sclerosis | circ_0005402 | Down | [60] | |
| circ_0035560 | [61] | |||
| Prion disease | CDR1as/ciR-7 | Up | [56,62] | |
| Psychiatry | Schizophrenia | hsa_circRNA_104597 | Down | [63] |
| Respiratory | Tuberculosis | hsa_circRNA_001937 | Up | [64] |
| hsa_circRNA_005086 | Up | [35] | ||
| hsa_circRNA_009024 | Up | |||
| hsa_circRNA_104964 | Down | |||
| hsa_circRNA_102101 | Down | |||
| hsa_circRNA_104296 | Down | |||
| Siliocosis | hsa_circ_0058493 | Up | [65] | |
| circRNA-012091 | Down | [66] | ||
| Pulmonary hypertension | circ-GSAP | Down | [67] | |
| circ_0068481 | Up | [68] | ||
| Acute respiratory distress syndrome | hsa_circRNA_101952 | Up | [69] | |
| hsa_circRNA_101523 | Up | |||
| hsa_circRNA_102927 | Down | |||
| hsa_circRNA_100562 | Down | |||
| hsa_circRNA_102034 | Down | |||
| Endocrine | Diabetes mellitus | CiRS-7 | Up | [70] |
| hsa_circ_0068087 | [71] | |||
| hsa_circ_0124636 | [71] | |||
| hsa_circ_0139110 | ||||
| hsa_circ_0054633 | ||||
| Diabetes/glucose | CDR1as/cirRS-7 | Up | [70,72] | |
| Homeostasis/CVD | circRNA-HIPK3 | Up | [73] | |
| circRNA-WDR77 | Up | |||
| circANKRD36 | Up | |||
| Diabetic cardiomyopathy | circRNA_000203 | Up | [74] | |
| circRNA_010567 | Up | [44] | ||
| hsa-circ-0076631 | Up | [75] | ||
| Diabetic nephropathy | circRNA_15698 | Up | [59,75,76] | |
| Gestational diabetes | circ_5824 | Down | [75] | |
| circ_3636 | Down | |||
| circ_0395 | Down | |||
| Diabetic retinopathy | circRNA-0005015 | Up | [75] | |
| circRNA-HIPK3 | Up | [77] | ||
| cZNF609 | Up | [78] | ||
| circRNA-cPWWP2A | Up | [79] |
| Cancer | circRNA | Expression | Function/target | Ref. |
| Lung cancer | hsa_circ_0005962 | Up | Promotes proliferation; non-invasive diagnostic biomarker | [80] |
| hsa_circ_0086414 | Down | Cancer progression; non-invasive diagnostic biomarker | [80] | |
| hsa_circ_0102537 | Down | Biomarker for diagnosis; fluid shear stress and PI3K-Akt signaling pathway | [81] | |
| hsa_circ_0000190 | Up | Tumorigenesis and immune evasion; facilitates tumorigenesis and immune evasion by upregulating the Expression of soluble PD-L1 in non-small-cell lung cancer | [82] | |
| CDR1as | Up | Enduring growth signaling; regulating miR-219a-5p/SOX5 axis | [83] | |
| hsa_circ_0001715 | Up | Distant metastasis; potential diagnostic and prognostic Biomarker | [84] | |
| circPTK2 | Down | Activating invasion and metastasis; inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ | [85] | |
| circ_0067934 | Up | Activating invasion and metastasis; regulating miR-1182/KLF8 axis and activating Wnt/β-catenin pathway | [86] | |
| hsa_circ_0012673 | Up | Cancer cell proliferation and invasion; via miR-320a/LIMK18521 axis | [87] | |
| F-circEA-2a | Up | Promotes cell migration and invasion | [88] | |
| F-circEA | Up | Promotes cell migration and invasion; potential diagnostic value | [89] | |
| circRNA_100876 | Up | Distant metastasis; potential prognostic biomarker and therapeutic target | [90] | |
| circRNA_ZEB1/hsa_circ_0023404 | Up | Promotes proliferation, migration, and invasion; regulating miR-217/ZEB1 axis | [91] | |
| CiRS-7 | Up | Increased proliferation, migration, and invasion, yet reduced apoptosis; targets NF-κB signaling | [92] | |
| HCC | has_circ_0001445 | Up | Regulates proliferation and migration; targets miR-942-5p/ALX4 axis | [93] |
| has_circ_0027089 | Up | Biomarker for diagnosis of hepatitis-related HCC | [94] | |
| has_circ_0016788 | Up | Evading growth inhibitors; targets miR-486/CDK4 pathway | [95] | |
| circ-SMARCA5 | Down | Inhibits cell proliferation and promotes apoptosis; potential prediction and monitoring biomarker for HCC | [96] | |
| circSMAD2 | Down | Activating invasion and metastasis; inhibits epithelial-mesenchymal transition by targeting miR-629 | [97] | |
| has_circ_0064428 | Down | Immune-associated prognostic biomarker for HCC patients | [98] | |
| has_circ_0009582; circ_0037120 | Up | Biomarker for diagnosis | [99] | |
| has_circ_0140117 | Up | Potential biomarkers for predicting occurrence of HCC | [99] | |
| CDR1as | Up | Cell proliferation and migration; enduring growth signaling | [100] | |
| Gastric cancer | has_circ_0010882 | Up | Role in proliferation, migration, and invasive phenotypes; regulation of PI3K/Akt/mTOR signaling | [101] |
| has_circ_0001017 | Down | Cell proliferation, migration, and invasion; sponge of miR-197 | [102] | |
| has_circ_0061276; hsa_circ_0001017 | Down | Potential prognostic tumor biomarkers | [103] | |
| has_circ_0000745 | Down | Regulating GC growth and migration; promising diagnostic biomarker | [104] | |
| circ-Rangap1 | Up | Cancer invasion and metastasis; targeting miR-877-3p | [105] | |
| has_circ_0000181 | Up | Distant metastasis and TNM stage; diagnostic biomarker | [106] | |
| circ-PSMC3 | Down | Cell proliferation and metastasis | [107] | |
| circ-LMTK2 | Down | |||
| circ-DLST | Up | |||
| circ-KIAA1244 | Down | TNM stage and lymphatic metastasis | [108] | |
| circ-ZFR | Down | Evading growth inhibitors; sponging miR-130a/miR-107 and modulating PTEN | [109] | |
| CDR1as | Down | Evading growth inhibitors; targeting miR-876-5p/GNG7 axis | [110] | |
| circRNA-000425 | Down | Evading growth inhibitors; proliferation, apoptosis, and cell drug sensitivity via YAP1-induced tumorigenesis | [111] | |
| circRNA_0023642 | Up | Activating invasion and metastasis; regulating epithelial-mesenchymal transition | [112] | |
| has_circ_0001178 | Up | Invasion and metastasis; via sponging multiple miRNAs | [113] | |
| has_circ_0005927 | Down | Cell colony-forming ability, migration, and invasion; regulating miR-942-5p/BATF2 axis | [114] | |
| has_circ_0082182 | Up | Drug resistance and cancer progression; sponging miR-326 | [115] | |
| hsa_circ_0082182, hsa_circ_0000370 | Up | Inhibit apoptosis; potential diagnostic markers | [116] | |
| has_circ_0035445 | Up | Promotes proliferation and migration but suppresses apoptosis; potential diagnostic marker | [116] | |
| has_circ_0006990 | Up | Cancer progression; via mediation of hsa_circ_0006990/miR-132-3p/MUC13 axis | [117] | |
| CDR1as | Up | Enduring growth signaling; regulating microRNA-7 | [118] | |
| circHIPK3 | Up | Enduring growth signaling; sponging miR-7 | [119] | |
| hsa_circ_0007534 | Up | Blocking apoptosis | [120] | |
| hsa_circRNA_103809 | Down | Cell proliferation and migration; via miR-532-3p/FOXO4 axis | [121] | |
| hsa-circ-0020397 | Up | Regulates CRC cell viability, apoptosis, and invasion by promoting the expression of miR-138 target genes | [121] | |
| Breast cancer | hsa_circ_0001785 | Up | Proliferation, migration, and invasion; potential diagnostic value | [122] |
| hsa_circ_0008673 | Up | Prognostic predictor of OS and DSS | [122] | |
| circ-ITCH | Down | Evading growth inhibitors; targets Wnt/β-catenin pathway | [123] | |
| hsa_circ_0104824 | Down | Cell migration, cell-cell adhesion, and proliferation; promising predictive biomarker and therapeutic target | [124] | |
| Esophageal cancer | circ-TTC17 | Up | Promotes proliferation and migration; novel biomarker for diagnosis, treatment, and prognosis | [125] |
| hsa_circ_0007203 (circ-DLG1) | Up | Promotes cell proliferation | [126] | |
| hsa_circ_0004771 | Up | Diagnostic biomarker; targets via miR-339-5p/CDC25A axis | [127] | |
| circGSK3β | Up | Promoting metastasis; augmenting β-catenin signaling | [128] | |
| circ-SLC7A5 | Up | Regulator of tumorigenesis and metastasis | [129] | |
| Pancreatic cancer | circ-LDLRAD3 | Up | Proliferation, migration and invasion; via miR-137-3p/PTN axis | [130,131] |
| circNFIB1, hsa_circ_0086375 | Down | Lymphangiogenesis and lymphatic metastasis; via miR-486-5p/PIK3R1/VEGF-C axis | [132] | |
| Thyroid cancer | has_circ_0124055 | Up | Regulates proliferation and apoptosis: Prognostic and diagnostic indicator | [133] |
| has_circ_0101622 | Up | Regulates proliferation and apoptosis: Prognostic and diagnostic indicator | [133] | |
| Gallbladder cancer | circ-MTO1 | Up | Early diagnostic and prognostic marker | [134] |
| circHIPK3 | Up | Evading growth inhibitors; sponging miR-124 | [135] | |
| Ovarian/endometrial cancer | has_circ_0078607 | Up | Adverse prognostic indicator for ovarian cancer; regulating miR-32-5p/SIK1 network | [136] |
| has_circ_0061140 | Up | Activating invasion and metastasis; targeting miR-197/high mobility group protein A1 axis in endometrial cancer | [137] | |
| Glioma/glioblastoma | hsa_circ_0046701 | Up | Enduring growth signaling; critical regulatory roles via hsa_circ_0046701/miR-142-3p/ITGB8 axis | [138] |
| circ-FBXW7 | Down | Enduring growth signaling; potential prognostic implications | [139] | |
| circNFIX | Up | Blocking apoptosis; regulating miR-378e/RPN2 axis | [140] | |
| cZNF292 | Up | Promoting angiogenesis; Wnt/β-catenin signaling | [141] |
Genome-wide investigations have found many circRNAs are conserved during evolution and enormous in number. CircRNAs have been categorized through extensive collections of the RNA sequencing data[14-16]. As circRNAs lack a poly(A) tail, the possible circRNA isoforms were identified via search for sequencing reads indicating a junction between two "scrambled" exons. A published landmark research article revealed that the junction sites of many circRNAs in rice (Oryza sativa) are flanked by diverse non-GT/AG splicing signals whereas most human exonic circRNAs are flanked by canonical GT/AG splicing signals[17]. This research provided a method for genome-wide identification of full-length circRNAs and increased the understanding of splicing signals of circRNAs. Most of the circRNAs functionally remains indefinable with only few exceptions. Table 3 summarizes the preliminary attempts of genome-wide circRNA identification in the human genome[18-20].
| Ref. | Genome-wide identification of circRNAs |
| Salzman et al[18], 2012 | Aimed to distinguish cancer-specific exon scrambling events |
| Identified 2748 scrambled isoforms in Hela and H9 embryonic stem cells | |
| Conclusion: 98% of scrambled isoforms represent circRNAs | |
| Jeck et al[8], 2013 | Classified circular transcripts based on their levels of abundance using three stringencies categories (low, medium, high) |
| Conclusion: circRNAs are conserved, stable, and nonrandom products of RNA splicing that could be involved in the control of gene expression | |
| Memczak et al[7], 2013 | Developed a computational method to detect circRNAs |
| Conclusion: circRNAs form a significant class of post-transcriptional regulators | |
| Guo et al[19], 2014 | Identified and quantified human circRNAs from ENCODE Ribozero RNA-seq data |
| Conclusion: Most circRNAs are nonsignificant side-products of splicing error | |
| Zhang et al[20], 2014 | Developed CIRCexplorer to distinguish thousands of circRNAs in humans with p(A)-wloRNase R RNA-seq data |
| Conclusion: Alternative circularization paired with alternative splicing can generate additional circRNAs from one gene, which is suggestive of the new line of complexity in gene regulation |
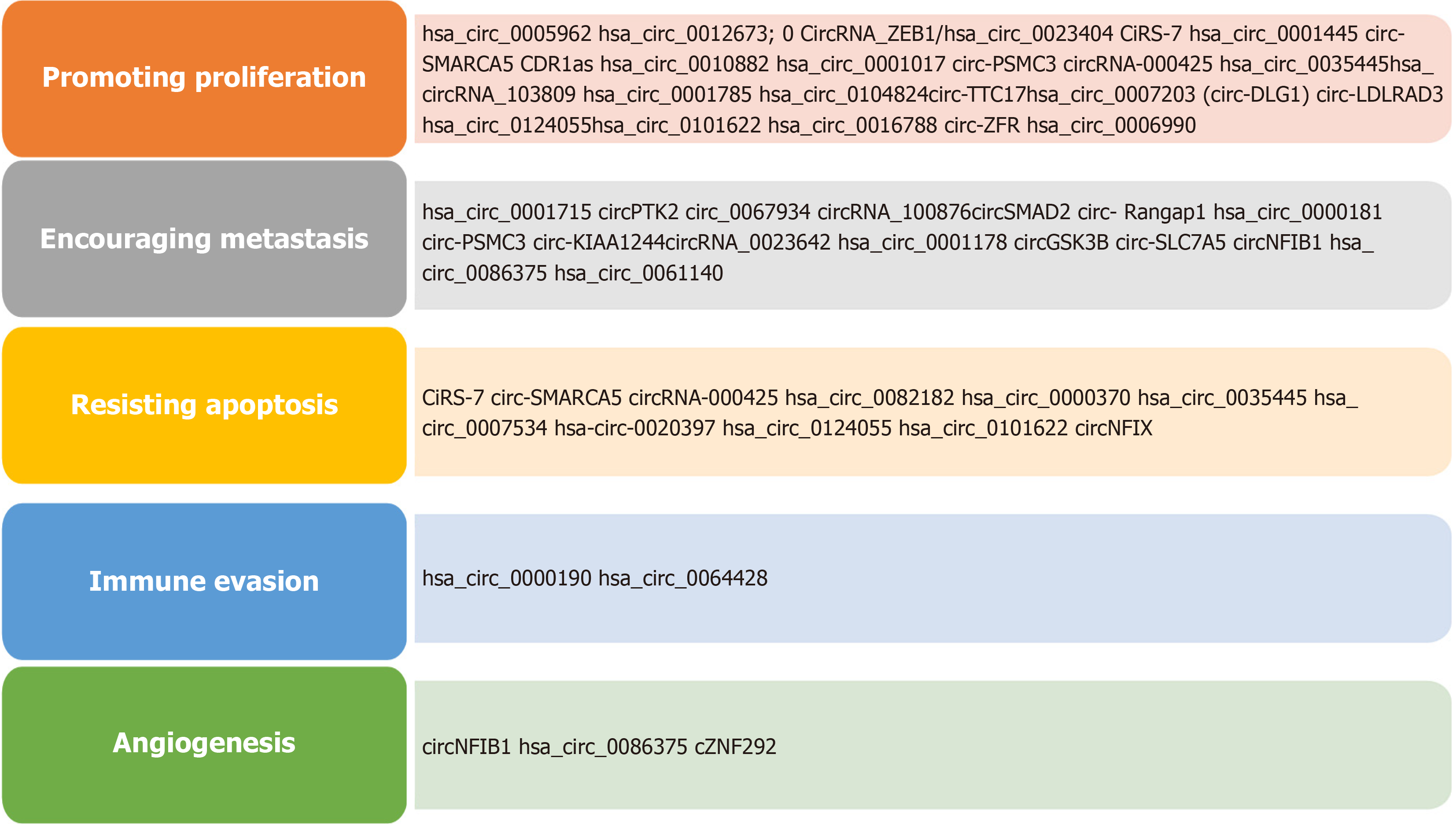
CircRNAs are extensively studied in cancers and there is huge data available showing their tissue-specific and cancer-specific expression patterns. Cancer-based studies revealed the clinical relevance of circRNAs like cancer-associated biomarkers, prognosis indicators, and formulation of treatment options. CircRNAs have been detected in liquid biopsies such as in various body fluids like plasma, blood, saliva, and urine making it an excellent choice as a non-invasive diagnosis for cancer[21]. There are many circRNAs identified in various cancers, which have diverse functions such as maintaining growth signaling, escaping growth inhibitors, resisting apoptosis, uncontrolled replicative immortality, promoting angiogenesis, and activating invasion and metastasis. The malignant cell must have one or more of these characteristics to gain immortality. CircRNAs are highly dysregulated in various human cancers and contribute to dif
The most intriguing circRNAs that have drawn the focus of scientists are the fusion-circRNAs (f-circRNA) which can arise from tumor-associated chromosomal translocations. F-circRNAs are capable of stimulating cellular transformation, therapeutic resistance, and tumor cell survival[22]. Previous studies identified circRNAs derived from cancer-associated chromosomal translocations and showed their tumor-promoting properties. These studies further found that tumor-associated chromosomal rearrangements lead to formation of f-circRNAs that are produced from transcribed exons of distinct genes affected by the translocations[23,24]. F-circRNAs participate in a variety of functions including cellular transformation and stimulating cell viability and/or resistance upon therapy[23,24]. Their work demonstrated the exis
CircRNAs levels are dynamically altered in neuronal cells throughout differentiation and many of them are enhanced in synapses. Moreover, there is evidence showing that circRNAs also accumulate with age, which is still a less explored area of biology[25]. Collectively, existing data indicate that circRNAs have crucial functions in synaptic plasticity and neuronal function. CircRNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed[26]. Numerous studies have shown that circRNAs are more enriched in neuronal tissues and are often originated from genes specific for neuronal and synaptic function. In addition, their expression is regulated during neuronal development by synaptic plasticity, indicating their potential in specific neuronal functions[27].
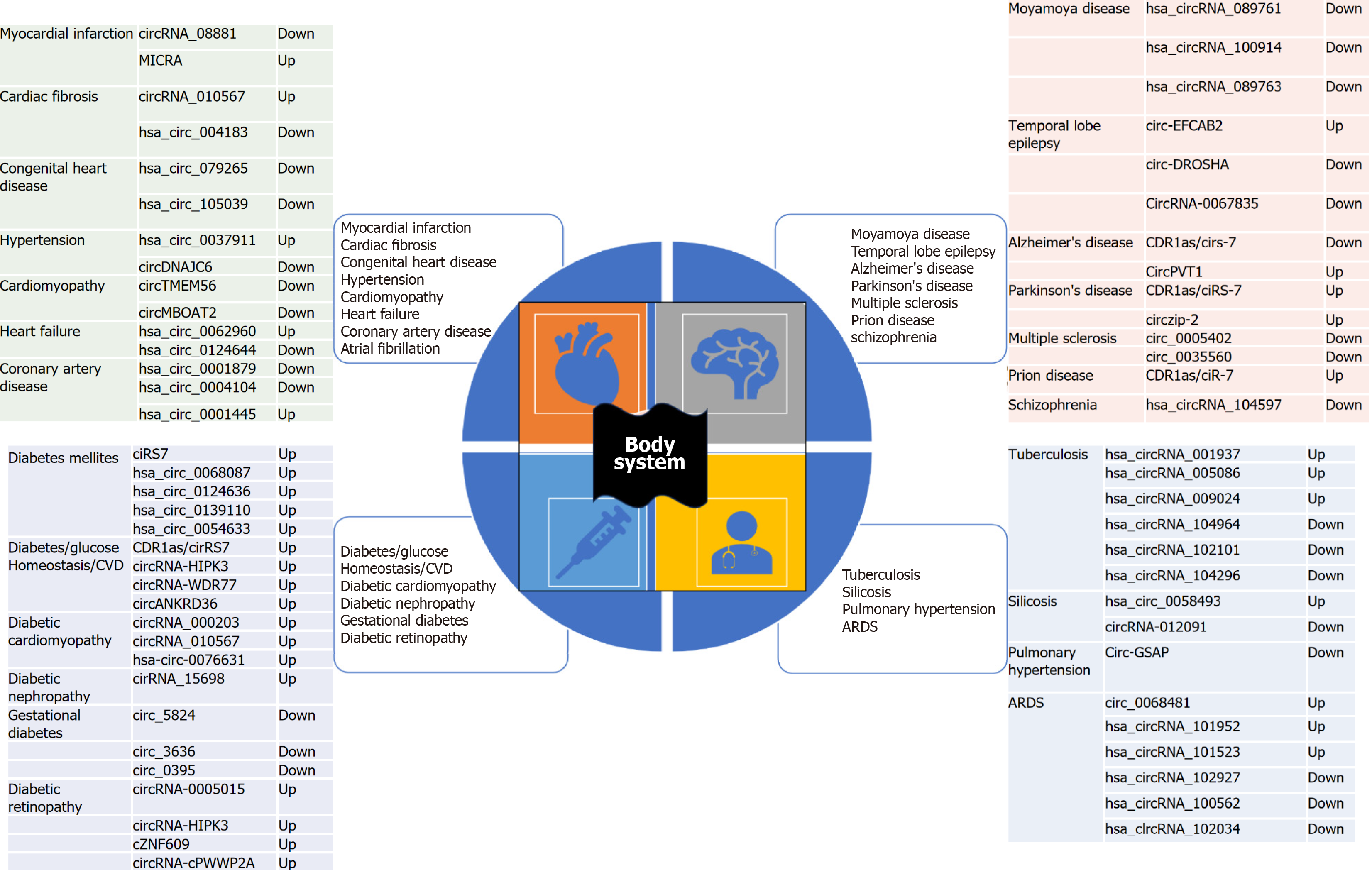
The involvement of circRNAs in various endocrine disorders like gynecological disease, Graves’ disease, age-related macular degeneration, and diabetes has been studied widely. However, this review only focuses on the landmark dis
A previous study observed that elevated glucose levels might modify circRNA expression in endothelial cells. This connection between endothelial cells and diabetic associated complications is mediated by circRNAs, which are the center of the events causing the pathogenesis of hyperglycemic endothelial injury[29]. Moreover, the ciRS-7–miR-7 axis is deeply linked to diabetes[30]. In support of the above statement, previous research data has shown that pancreatic islet cells highly express miR-7 that ultimately blocks beta-cell proliferation, leading to disrupted rapamycin (mTOR) signaling pa
CircRNAs are broadly expressed in mammalian cells, and it they have a crucial role in cardiogenesis. The literature shows that 1702 circRNAs are expressed during cardiogenesis[32], which contribute to cell specification and differentiation[33,34]. Further evidence suggests that circRNAs have a crucial role in the pathogenesis of cardiac disorders[35]. Hence, these can act as potential biomarkers in cardiovascular diseases.
Heart-related circRNA (HRCR) was the first circRNA found to be inhibited in hypertrophic hearts. Researchers noticed that HRCR binds and impedes the miRNA miR-223, which was associated with cardiac hypertrophy[36]. Additionally, it has also been found that the circRNA CDR1AS binds to miR-7 and miR-7a and exacerbates myocardial infarction-mediated cardiomyocytes loss[37]. CircRNA_081881 expression levels drop by 10-times in myocardial infarction patients’ blood, which makes this circRNA a potential biomarker for cardiac diseases[38]. Although the evidence present in the literature are intriguing and exciting, additional research on circRNAs using advanced technologies can enlighten this relatively less explored area of research.
CircRNAs are implicated in malignant and non-malignant respiratory disorders. Many research articles are suggestive of their prospective as biomarkers for respiratory disorders. To date, circRNAs have been identified in lung cancer as well as in non-cancerous respiratory diseases like pulmonary hypertension, pulmonary tuberculosis (TB), acute respiratory distress syndrome, and silicosis, but they may not be limited to just these entries mentioned here[39]. One of the exciting roles of circRNAs which made a headline previously is the diagnosis of TB. A study by Fu et al[40] confirmed that blood samples could be used to diagnose TB. They examined circRNAs in peripheral blood mononuclear cells in healthy individuals and TB patients regarding the diagnosis of TB using circRNAs[40]. Hence, circRNAs can act as biomarkers to diagnose TB. In a nutshell, circRNA expression signatures could be probable biomarkers in the diagnosis and prognosis of various respiratory disorders.
Figure 4 shows important roles of circRNAs in various non-malignant human diseases. Figure 5 shows summarized view of circRNAs as potential biomarkers in major human cancers classification based on cancer hallmark signatures[41,42].
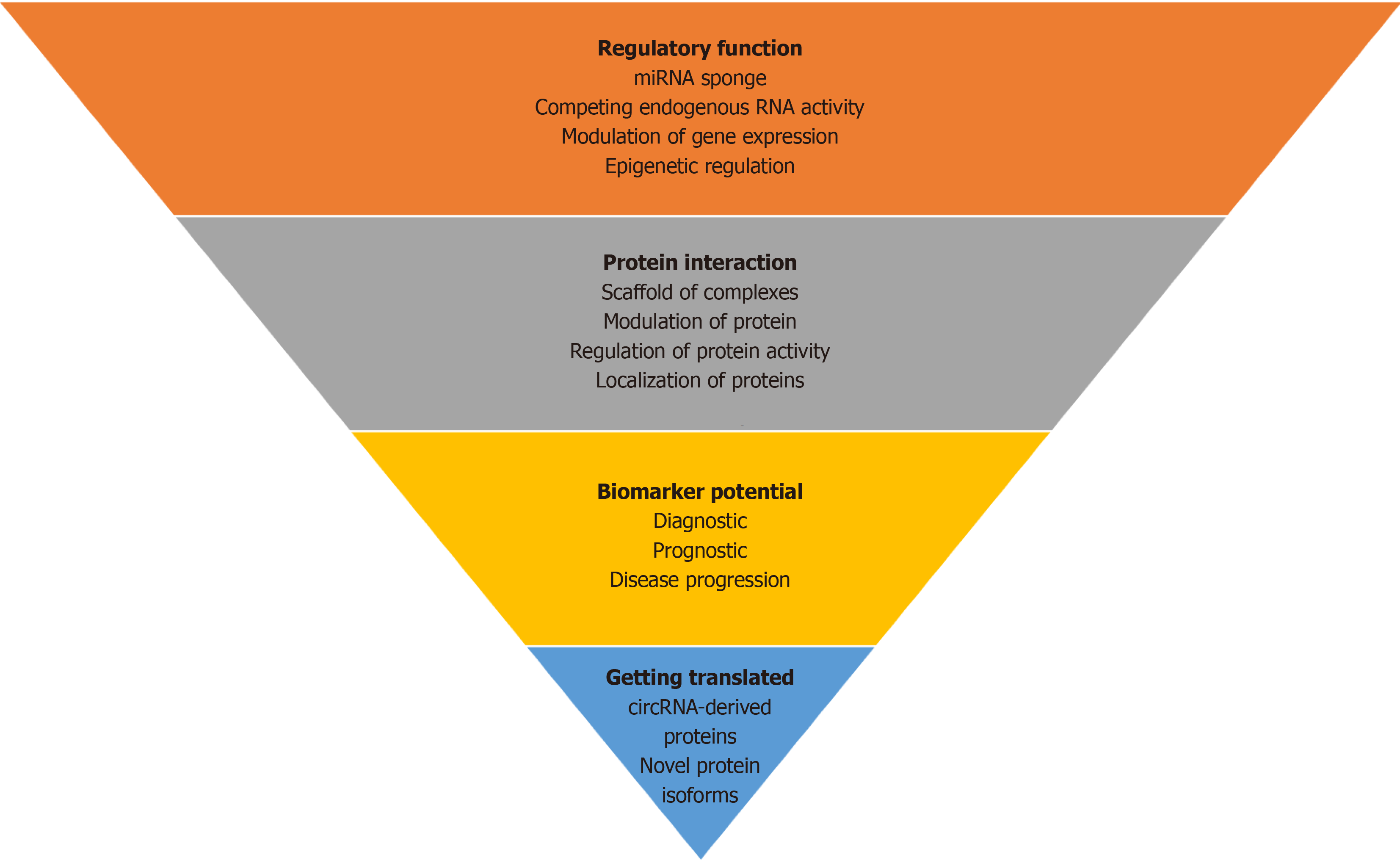
The emerging evidence from experimental settings has shown the crucial role of circRNAs in various human diseases. A single circRNA can perform multiple functions including but not limited to miRNA sponging, protein sponging, translation, spliceosome regulation, and epigenetic regulation. Therefore, targeting circRNAs to manipulate the effect of its downstream targets like miRNAs and genes can prove to be a new powerful treatment-oriented approach for multiple diseases. The covalently closed loop like structure of circRNAs is highly stable compared to other RNAs and can be detected in body fluids like saliva, blood, and urine. These characteristics make circRNAs an favorable choice for bio
However, circRNAs are still underrepresented targets in molecular biology because of the lack of enough functional studies. A few challenges associated with circRNAs are that they are not detected in RNA sequencing data because of the lack of poly(A) tail and hence easily ignored from most of RNA sequencing studies. Moreover, circRNAs perform a diverse range of complex functions that require a high level of computational skills to decode. The current methods of circRNA detection in body fluids have limitations and are associated with high cost. These challenges can be solved with further advancement of research by future studies. Although circRNAs are favorable disease biomarkers, there are yet various key problems to be addressed for the application of circRNAs in disease settings and further evidence is needed to support the clinical significance of circRNAs in specific diseases.
CircRNAs play critical roles in numerous diseases. Once thought to be functionless, circRNAs have now become a major research agenda. Most of the research articles enlisted in this review are related to the relationship between the expre
| 1. | Ren S, Lin P, Wang J, Yu H, Lv T, Sun L, Du G. Circular RNAs: Promising Molecular Biomarkers of Human Aging-Related Diseases via Functioning as a miRNA Sponge. Mol Ther Methods Clin Dev. 2020;18:215-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 2. | Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993-1997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 181] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 3. | Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 713] [Cited by in RCA: 767] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 4. | Zhang Y, Li J, Chen R, Dai A, Luan D, Ma T, Hua D, Chen G, Chang G. Cloning, characterization and widespread expression analysis of testicular piRNA-like chicken RNAs. Mol Biol Rep. 2013;40:2799-2807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 5. | Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 675] [Cited by in RCA: 780] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 6. | Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 758] [Cited by in RCA: 950] [Article Influence: 28.8] [Reference Citation Analysis (1)] |
| 7. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6229] [Article Influence: 479.2] [Reference Citation Analysis (0)] |
| 8. | Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3481] [Cited by in RCA: 3538] [Article Influence: 272.2] [Reference Citation Analysis (0)] |
| 9. | Wang Z. Not just a sponge: new functions of circular RNAs discovered. Sci China Life Sci. 2015;58:407-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Mo Y, Liu Y, Lu A, Zhang H, Tang L. Role of circRNAs in viral infection and their significance for diagnosis and treatment (Review). Int J Mol Med. 2021;47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 11. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6225] [Article Influence: 478.8] [Reference Citation Analysis (0)] |
| 12. | Dou C, Cao Z, Yang B, Ding N, Hou T, Luo F, Kang F, Li J, Yang X, Jiang H, Xiang J, Quan H, Xu J, Dong S. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci Rep. 2016;6:21499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 152] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 13. | Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 553] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 14. | Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang J, Cheng H, Yan J, Zhang S, Yang P, Zhao F. Expanded Expression Landscape and Prioritization of Circular RNAs in Mammals. Cell Rep. 2019;26:3444-3460.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 211] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 15. | Cheng J, Metge F, Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2016;32:1094-1096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 278] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 16. | Gaffo E, Buratin A, Dal Molin A, Bortoluzzi S. Bioinformatic Analysis of Circular RNA Expression. Methods Mol Biol. 2021;2348:343-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 17. | Ye CY, Zhang X, Chu Q, Liu C, Yu Y, Jiang W, Zhu QH, Fan L, Guo L. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol. 2017;14:1055-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 95] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 18. | Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1589] [Cited by in RCA: 2017] [Article Influence: 144.1] [Reference Citation Analysis (0)] |
| 19. | Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1021] [Cited by in RCA: 1308] [Article Influence: 109.0] [Reference Citation Analysis (0)] |
| 20. | Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1186] [Cited by in RCA: 1545] [Article Influence: 128.8] [Reference Citation Analysis (0)] |
| 21. | Peter MR, Zhao F, Jeyapala R, Kamdar S, Xu W, Hawkins C, Evans AJ, Fleshner NE, Finelli A, Bapat B. Investigating Urinary Circular RNA Biomarkers for Improved Detection of Renal Cell Carcinoma. Front Oncol. 2021;11:814228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Oncogenic Circular RNAs Arise from Chromosomal Translocations. Cancer Discov. 2016;6:OF20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;166:1055-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 155] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 24. | Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;165:289-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 484] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 25. | Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol. 2017;14:1028-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 189] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 26. | Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1370] [Cited by in RCA: 1876] [Article Influence: 170.5] [Reference Citation Analysis (0)] |
| 27. | Chen W, Schuman E. Circular RNAs in Brain and Other Tissues: A Functional Enigma. Trends Neurosci. 2016;39:597-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 149] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 28. | Tu C, Wang L, Wei L, Jiang Z. The role of circular RNA in Diabetic Nephropathy. Int J Med Sci. 2022;19:916-923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Wei H, Cao C, Wei X, Meng M, Wu B, Meng L, Wei X, Gu S, Li H. Circular RNA circVEGFC accelerates high glucose-induced vascular endothelial cells apoptosis through miR-338-3p/HIF-1α/VEGFA axis. Aging (Albany NY). 2020;12:14365-14375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 30. | Agbu P, Cassidy JJ, Braverman J, Jacobson A, Carthew RW. MicroRNA miR-7 Regulates Secretion of Insulin-Like Peptides. Endocrinology. 2020;161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes. 2013;62:887-895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 163] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 32. | Li Y, Zhang J, Huo C, Ding N, Li J, Xiao J, Lin X, Cai B, Zhang Y, Xu J. Dynamic Organization of lncRNA and Circular RNA Regulators Collectively Controlled Cardiac Differentiation in Humans. EBioMedicine. 2017;24:137-146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 33. | Luo JY, Zhang Y, Wang L, Huang Y. Regulators and effectors of bone morphogenetic protein signalling in the cardiovascular system. J Physiol. 2015;593:2995-3011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | E S, Costa MC, Kurc S, Drożdż A, Cortez-Dias N, Enguita FJ. The circulating non-coding RNA landscape for biomarker research: lessons and prospects from cardiovascular diseases. Acta Pharmacol Sin. 2018;39:1085-1099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 35. | Lee ECS, Elhassan SAM, Lim GPL, Kok WH, Tan SW, Leong EN, Tan SH, Chan EWL, Bhattamisra SK, Rajendran R, Candasamy M. The roles of circular RNAs in human development and diseases. Biomed Pharmacother. 2019;111:198-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 36. | Wu S, Chen L, Zhou X. Circular RNAs in the regulation of cardiac hypertrophy. Mol Ther Nucleic Acids. 2022;27:484-490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS One. 2016;11:e0151753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 330] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 38. | Wang W, Wang Y, Piao H, Li B, Huang M, Zhu Z, Li D, Wang T, Xu R, Liu K. Circular RNAs as potential biomarkers and therapeutics for cardiovascular disease. PeerJ. 2019;7:e6831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 39. | Wang J, Zhu M, Pan J, Chen C, Xia S, Song Y. Circular RNAs: a rising star in respiratory diseases. Respir Res. 2019;20:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 40. | Fu Y, Wang J, Qiao J, Yi Z. Signature of circular RNAs in peripheral blood mononuclear cells from patients with active tuberculosis. J Cell Mol Med. 2019;23:1917-1925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 41. | Najafi S. Circular RNAs as emerging players in cervical cancer tumorigenesis; A review to roles and biomarker potentials. Int J Biol Macromol. 2022;206:939-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 70] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 42. | Najafi S. The emerging roles and potential applications of circular RNAs in ovarian cancer: a comprehensive review. J Cancer Res Clin Oncol. 2023;149:2211-2234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 43. | Siede D, Rapti K, Gorska AA, Katus HA, Altmüller J, Boeckel JN, Meder B, Maack C, Völkers M, Müller OJ, Backs J, Dieterich C. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol. 2017;109:48-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 103] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 44. | Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun. 2017;487:769-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 305] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 45. | Wu J, Li J, Liu H, Yin J, Zhang M, Yu Z, Miao H. Circulating plasma circular RNAs as novel diagnostic biomarkers for congenital heart disease in children. J Clin Lab Anal. 2019;33:e22998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 46. | Bao X, Zheng S, Mao S, Gu T, Liu S, Sun J, Zhang L. A potential risk factor of essential hypertension in case-control study: Circular RNA hsa_circ_0037911. Biochem Biophys Res Commun. 2018;498:789-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 47. | Sonnenschein K, Wilczek AL, de Gonzalo-Calvo D, Pfanne A, Derda AA, Zwadlo C, Bavendiek U, Bauersachs J, Fiedler J, Thum T. Serum circular RNAs act as blood-based biomarkers for hypertrophic obstructive cardiomyopathy. Sci Rep. 2019;9:20350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 48. | Sun Y, Jiang X, Lv Y, Liang X, Zhao B, Bian W, Zhang D, Jiang J, Zhang C. Circular RNA Expression Profiles in Plasma from Patients with Heart Failure Related to Platelet Activity. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 49. | Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:39918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 191] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 50. | Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu X, Li L, Yang B, Chen J, Chen S, Lu X, Gu D. Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis. 2019;286:88-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 51. | Vilades D, Martínez-Camblor P, Ferrero-Gregori A, Bär C, Lu D, Xiao K, Vea À, Nasarre L, Sanchez Vega J, Leta R, Carreras F, Thum T, Llorente-Cortés V, de Gonzalo-Calvo D. Plasma circular RNA hsa_circ_0001445 and coronary artery disease: Performance as a biomarker. FASEB J. 2020;34:4403-4414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 52. | Reckman YJ, Creemers EE. Circulating Circles Predict Postoperative Atrial Fibrillation. J Am Heart Assoc. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 53. | Zhao M, Gao F, Zhang D, Wang S, Zhang Y, Wang R, Zhao J. Altered expression of circular RNAs in Moyamoya disease. J Neurol Sci. 2017;381:25-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 54. | Li J, Lin H, Sun Z, Kong G, Yan X, Wang Y, Wang X, Wen Y, Liu X, Zheng H, Jia M, Shi Z, Xu R, Yang S, Yuan F. High-Throughput Data of Circular RNA Profiles in Human Temporal Cortex Tissue Reveals Novel Insights into Temporal Lobe Epilepsy. Cell Physiol Biochem. 2018;45:677-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 55. | Gong GH, An FM, Wang Y, Bian M, Wang D, Wei CX. Comprehensive Circular RNA Profiling Reveals the Regulatory Role of the CircRNA-0067835/miR-155 Pathway in Temporal Lobe Epilepsy. Cell Physiol Biochem. 2018;51:1399-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 56. | Akhter R. Circular RNA and Alzheimer's Disease. Adv Exp Med Biol. 2018;1087:239-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 177] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 57. | Najafi S, Aghaei Zarch SM, Majidpoor J, Pordel S, Aghamiri S, Fatih Rasul M, Asemani Y, Vakili O, Mohammadi V, Movahedpour A, Arghiani N. Recent insights into the roles of circular RNAs in human brain development and neurologic diseases. Int J Biol Macromol. 2023;225:1038-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 43] [Reference Citation Analysis (0)] |
| 58. | Ma Y, Liu Y, Jiang Z. CircRNAs: A new perspective of biomarkers in the nervous system. Biomed Pharmacother. 2020;128:110251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 59. | Ma C, Luan J, Kopp JB, Zhou H. Emerging Role of Circular RNAs in Kidney Diseases in Nephrology. Curr Drug Targets. 2022;23:330-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 60. | Zurawska AE, Mycko MP, Selmaj I, Raine CS, Selmaj KW. Multiple Sclerosis: circRNA Profile Defined Reveals Links to B-Cell Function. Neurol Neuroimmunol Neuroinflamm. 2021;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 61. | Gu J, Su C, Huang F, Zhao Y, Li J. Past, Present and Future: The Relationship Between Circular RNA and Immunity. Front Immunol. 2022;13:894707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 62. | Shao Y, Chen Y. Roles of Circular RNAs in Neurologic Disease. Front Mol Neurosci. 2016;9:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 63. | Yao G, Niu W, Zhu X, He M, Kong L, Chen S, Zhang L, Cheng Z. hsa_circRNA_104597: a novel potential diagnostic and therapeutic biomarker for schizophrenia. Biomark Med. 2019;13:331-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 64. | Huang ZK, Yao FY, Xu JQ, Deng Z, Su RG, Peng YP, Luo Q, Li JM. Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Active Tuberculosis Patients. Cell Physiol Biochem. 2018;45:1230-1240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 65. | Cheng Z, Zhang Y, Wu S, Zhao R, Yu Y, Zhou Y, Zhou Z, Dong Y, Qiu A, Xu H, Liu Y, Zhang W, Tian T, Wu Q, Gu H, Chu M. Peripheral blood circular RNA hsa_circ_0058493 as a potential novel biomarker for silicosis and idiopathic pulmonary fibrosis. Ecotoxicol Environ Saf. 2022;236:113451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 66. | Cheng Y, Luo W, Li Z, Cao M, Zhu Z, Han C, Dai X, Zhang W, Wang J, Yao H, Chao J. CircRNA-012091/PPP1R13B-mediated Lung Fibrotic Response in Silicosis via Endoplasmic Reticulum Stress and Autophagy. Am J Respir Cell Mol Biol. 2019;61:380-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 67. | Sun X, Chen R, Yao X, Zheng Z, Wang C, Chen J, Cheng J. circGSAP: A New Clinical Biomarker for Idiopathic Pulmonary Hypertension? Am J Respir Crit Care Med. 2022;205:252-253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 68. | Zhang Y, Chen Y, Yao H, Lie Z, Chen G, Tan H, Zhou Y. Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm Circ. 2019;9:2045894019888416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 69. | Jiang Y, Zhu F, Wu GS, Wang KA, Wang C, Yu Q, Zhu BH, Sun Y, Xia ZF. Microarray and bioinformatics analysis of circular RNAs expression profile in traumatic lung injury. Exp Ther Med. 2020;20:227-234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 70. | Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 324] [Cited by in RCA: 412] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 71. | Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237-245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 194] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 72. | Yin W, Zhang Z, Xiao Z, Li X, Luo S, Zhou Z. Circular RNAs in diabetes and its complications: Current knowledge and future prospects. Front Genet. 2022;13:1006307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 73. | Fang Y, Wang X, Li W, Han J, Jin J, Su F, Zhang J, Huang W, Xiao F, Pan Q, Zou L. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med. 2018;42:1865-1874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 90] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 74. | Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan R, Yuan SJ, Zeng N, Yang ZZ, Yang H, Wang XP, Duan JZ, Wang S, Luo JF, Wu SL, Shan ZX. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res. 2020;116:1323-1334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 206] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 75. | Zaiou M. circRNAs Signature as Potential Diagnostic and Prognostic Biomarker for Diabetes Mellitus and Related Cardiovascular Complications. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 76. | Jin J, Sun H, Shi C, Yang H, Wu Y, Li W, Dong YH, Cai L, Meng XM. Circular RNA in renal diseases. J Cell Mol Med. 2020;24:6523-6533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 120] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 77. | Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, Zhang YY, Liu B, Zhang SJ, Wang JJ, Zhang SH, Wu JH, Zhao C, Yan B. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation. 2017;136:1629-1642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 414] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 78. | Liu C, Yao MD, Li CP, Shan K, Yang H, Wang JJ, Liu B, Li XM, Yao J, Jiang Q, Yan B. Silencing Of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics. 2017;7:2863-2877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 225] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 79. | Yan Q, He X, Kuang G, Ou C. CircRNA cPWWP2A: an emerging player in diabetes mellitus. J Cell Commun Signal. 2020;14:351-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 80. | Liu XX, Yang YE, Liu X, Zhang MY, Li R, Yin YH, Qu YQ. A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J Transl Med. 2019;17:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 81. | Lin S, Xiong W, Liu H, Pei L, Yi H, Guan Y. Profiling and Integrated Analysis of Differentially Expressed Circular RNAs in Plasma Exosomes as Novel Biomarkers for Advanced-Stage Lung Adenocarcinoma. Onco Targets Ther. 2020;13:12965-12977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 82. | Luo YH, Yang YP, Chien CS, Yarmishyn AA, Adekunle Ishola A, Chien Y, Chen YM, Tsai PH, Lin TW, Wang ML, Chiou SH. Circular RNA hsa_circ_0000190 Facilitates the Tumorigenesis and Immune Evasion by Upregulating the Expression of Soluble PD-L1 in Non-Small-Cell Lung Cancer. Int J Mol Sci. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 83. | Li Y, Zhang J, Pan S, Zhou J, Diao X, Liu S. CircRNA CDR1as knockdown inhibits progression of non-small-cell lung cancer by regulating miR-219a-5p/SOX5 axis. Thorac Cancer. 2020;11:537-548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 84. | Lu GJ, Cui J, Qian Q, Hou ZB, Xie HY, Hu W, Hao KK, Xia N, Zhang Y. Overexpression of hsa_circ_0001715 is a Potential Diagnostic and Prognostic Biomarker in Lung Adenocarcinoma. Onco Targets Ther. 2020;13:10775-10783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 85. | Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, Liu Z, Zeng Y, Li C, Zhao J, Su Z, Zhang C, Liu X, Xu G, Zhang HT. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17:140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 290] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 86. | Zhao M, Ma W, Ma C. Circ_0067934 promotes non-small cell lung cancer development by regulating miR-1182/KLF8 axis and activating Wnt/β-catenin pathway. Biomed Pharmacother. 2020;129:110461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 87. | Qin H, Liu J, Du ZH, Hu R, Yu YK, Wang QA. Circular RNA hsa_circ_0012673 facilitates lung cancer cell proliferation and invasion via miR-320a/LIMK18521 axis. Eur Rev Med Pharmacol Sci. 2020;24:1841-1852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 88. | Liu J, Zhang X, Yan M, Li H. Emerging Role of Circular RNAs in Cancer. Front Oncol. 2020;10:663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 89. | Tan S, Sun D, Pu W, Gou Q, Guo C, Gong Y, Li J, Wei YQ, Liu L, Zhao Y, Peng Y. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer. 2018;17:138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 90. | Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, Nan KJ. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 206] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 91. | Liu C, Zhang Z, Qi D. Circular RNA hsa_circ_0023404 promotes proliferation, migration and invasion in non-small cell lung cancer by regulating miR-217/ZEB1 axis. Onco Targets Ther. 2019;12:6181-6189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 92. | Su C, Han Y, Zhang H, Li Y, Yi L, Wang X, Zhou S, Yu D, Song X, Xiao N, Cao X, Liu Z. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22:3097-3107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 93. | Xu Q, Zhou L, Yang G, Meng F, Wan Y, Wang L, Zhang L. Overexpression of circ_0001445 decelerates hepatocellular carcinoma progression by regulating miR-942-5p/ALX4 axis. Biotechnol Lett. 2020;42:2735-2747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 94. | Zhu K, Zhan H, Peng Y, Yang L, Gao Q, Jia H, Dai Z, Tang Z, Fan J, Zhou J. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. 2020;41:296-302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 95. | Guan Z, Tan J, Gao W, Li X, Yang Y, Li X, Li Y, Wang Q. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol. 2018;234:500-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 96. | Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, Deng Q, Zheng F. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta. 2019;492:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 97. | Zhang X, Luo P, Jing W, Zhou H, Liang C, Tu J. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther. 2018;11:2853-2863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 98. | Weng Q, Chen M, Li M, Zheng YF, Shao G, Fan W, Xu XM, Ji J. Global microarray profiling identified hsa_circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet. 2019;56:32-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 99. | Wu C, Deng L, Zhuo H, Chen X, Tan Z, Han S, Tang J, Qian X, Yao A. Circulating circRNA predicting the occurrence of hepatocellular carcinoma in patients with HBV infection. J Cell Mol Med. 2020;24:10216-10222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 100. | Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, Qi F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 2019;11:8183-8203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 101. | Peng YK, Pu K, Su HX, Zhang J, Zheng Y, Ji R, Guo QH, Wang YP, Guan QL, Zhou YN. Circular RNA hsa_circ_0010882 promotes the progression of gastric cancer via regulation of the PI3K/Akt/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24:1142-1151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 102. | Li H, Shan C, Wang J, Hu C. CircRNA Hsa_circ_0001017 Inhibited Gastric Cancer Progression via Acting as a Sponge of miR-197. Dig Dis Sci. 2021;66:2261-2271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 103. | Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96:85-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 216] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 104. | Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23:6330-6338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 133] [Cited by in RCA: 178] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 105. | Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, Zheng CH, Li P, Huang CM. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 199] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 106. | Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32:e22333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 107. | Ruan Y, Li Z, Shen Y, Li T, Zhang H, Guo J. Functions of circular RNAs and their potential applications in gastric cancer. Expert Rev Gastroenterol Hepatol. 2020;14:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 108. | Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17:137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 206] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 109. | Liu T, Liu S, Xu Y, Shu R, Wang F, Chen C, Zeng Y, Luo H. Circular RNA-ZFR Inhibited Cell Proliferation and Promoted Apoptosis in Gastric Cancer by Sponging miR-130a/miR-107 and Modulating PTEN. Cancer Res Treat. 2018;50:1396-1417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 110. | Jiang J, Li R, Wang J, Hou J, Qian H, Xu W. Circular RNA CDR1as Inhibits the Metastasis of Gastric Cancer through Targeting miR-876-5p/GNG7 Axis. Gastroenterol Res Pract. 2021;2021:5583029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 111. | Liu Z, Huang S, Cao Y, Yao Y, Li J, Chen J, Jiang B, Yuan X, Xiang X, Xiong J, Deng J. YAP1 inhibits circRNA-000425 expression and thus promotes oncogenic activities of miR-17 and miR-106. Biochem Biophys Res Commun. 2018;503:2370-2375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 112. | Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22:2297-2303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 48] [Reference Citation Analysis (0)] |
| 113. | Ren C, Zhang Z, Wang S, Zhu W, Zheng P, Wang W. Circular RNA hsa_circ_0001178 facilitates the invasion and metastasis of colorectal cancer through upregulating ZEB1 via sponging multiple miRNAs. Biol Chem. 2020;401:487-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 114. | Yu C, Li D, Yan Q, Wang Y, Yang X, Zhang S, Zhang Y, Zhang Z. Circ_0005927 Inhibits the Progression of Colorectal Cancer by Regulating miR-942-5p/BATF2 Axis. Cancer Manag Res. 2021;13:2295-2306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 115. | Wang Z, Liu J, Yang T, Wang Q, Liang R, Tang J. Circ_0082182 upregulates the NFIB level via sponging miR-326 to promote oxaliplatin resistance and malignant progression of colorectal cancer cells. Mol Cell Biochem. 2023;478:1045-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 116. | Ye DX, Wang SS, Huang Y, Chi P. A 3-circular RNA signature as a noninvasive biomarker for diagnosis of colorectal cancer. Cancer Cell Int. 2019;19:276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 117. | Chen B, Wu L, Tang X, Wang T, Wang S, Yu H, Wan G, Xie M, Zhang R, Xiao H, Deng W. Quercetin Inhibits Tumorigenesis of Colorectal Cancer Through Downregulation of hsa_circ_0006990. Front Pharmacol. 2022;13:874696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 118. | Tang W, Ji M, He G, Yang L, Niu Z, Jian M, Wei Y, Ren L, Xu J. Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. Onco Targets Ther. 2017;10:2045-2056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 138] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 119. | Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 447] [Cited by in RCA: 492] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 120. | Zhang R, Xu J, Zhao J, Wang X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 41] [Reference Citation Analysis (0)] |
| 121. | Bian L, Zhi X, Ma L, Zhang J, Chen P, Sun S, Li J, Sun Y, Qin J. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p / FOXO4 axis. Biochem Biophys Res Commun. 2018;505:346-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 142] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 122. | Yin WB, Yan MG, Fang X, Guo JJ, Xiong W, Zhang RP. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 139] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 123. | Wang ST, Liu LB, Li XM, Wang YF, Xie PJ, Li Q, Wang R, Wei Q, Kang YH, Meng R, Feng XH. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/β-catenin pathway. Neoplasma. 2019;66:232-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 124. | Li X, Ma F, Wu L, Zhang X, Tian J, Li J, Cao J, Ma Y, Zhang L, Wang L. Identification of Hsa_circ_0104824 as a Potential Biomarkers for Breast Cancer. Technol Cancer Res Treat. 2020;19:1533033820960745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 125. | Wang Q, Zhang Q, Sun H, Tang W, Yang L, Xu Z, Liu Z, Jin H, Cao X. Circ-TTC17 Promotes Proliferation and Migration of Esophageal Squamous Cell Carcinoma. Dig Dis Sci. 2019;64:751-758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 126. | Rong J, Wang Q, Zhang Y, Zhu D, Sun H, Tang W, Wang R, Shi W, Cao XF. Circ-DLG1 promotes the proliferation of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11:6723-6730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 127. | Huang E, Fu J, Yu Q, Xie P, Yang Z, Ji H, Wang L, Luo G, Zhang Y, Li K. CircRNA hsa_circ_0004771 promotes esophageal squamous cell cancer progression via miR-339-5p/CDC25A axis. Epigenomics. 2020;12:587-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 128. | Hu X, Wu D, He X, Zhao H, He Z, Lin J, Wang K, Wang W, Pan Z, Lin H, Wang M. circGSK3β promotes metastasis in esophageal squamous cell carcinoma by augmenting β-catenin signaling. Mol Cancer. 2019;18:160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 142] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 129. | Wang Q, Liu H, Liu Z, Yang L, Zhou J, Cao X, Sun H. Circ-SLC7A5, a potential prognostic circulating biomarker for detection of ESCC. Cancer Genet. 2020;240:33-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 130. | Yao J, Zhang C, Chen Y, Gao S. Downregulation of circular RNA circ-LDLRAD3 suppresses pancreatic cancer progression through miR-137-3p/PTN axis. Life Sci. 2019;239:116871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (1)] |
| 131. | Sayad A, Najafi S, Hussen BM, Jamali E, Taheri M, Ghafouri-Fard S. The role of circular RNAs in pancreatic cancer: new players in tumorigenesis and potential biomarkers. Pathol Res Pract. 2022;232:153833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 132. | Kong Y, Li Y, Luo Y, Zhu J, Zheng H, Gao B, Guo X, Li Z, Chen R, Chen C. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol Cancer. 2020;19:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 139] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 133. | Sun JW, Qiu S, Yang JY, Chen X, Li HX. Hsa_circ_0124055 and hsa_circ_0101622 regulate proliferation and apoptosis in thyroid cancer and serve as prognostic and diagnostic indicators. Eur Rev Med Pharmacol Sci. 2020;24:4348-4360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 134. | Ghafouri-Fard S, Khoshbakht T, Bahranian A, Taheri M, Hallajnejad M. CircMTO1: A circular RNA with roles in the carcinogenesis. Biomed Pharmacother. 2021;142:112025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 135. | Kai D, Yannian L, Yitian C, Dinghao G, Xin Z, Wu J. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem Biophys Res Commun. 2018;503:863-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 82] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 136. | Jin Y, Wang H. Circ_0078607 inhibits the progression of ovarian cancer via regulating the miR-32-5p/SIK1 network. J Ovarian Res. 2022;15:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 137. | Zhou Y, Pan A, Zhang Y, Li X. Hsa_circ_0039569 facilitates the progression of endometrial carcinoma by targeting the miR-197/high mobility group protein A1 axis. Bioengineered. 2022;13:4212-4225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 138. | Li G, Yang H, Han K, Zhu D, Lun P, Zhao Y. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem Biophys Res Commun. 2018;498:254-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 139. | Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst. 2018;110:304-315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 858] [Cited by in RCA: 857] [Article Influence: 107.1] [Reference Citation Analysis (0)] |
| 140. | Ding C, Wu Z, You H, Ge H, Zheng S, Lin Y, Wu X, Lin Z, Kang D. CircNFIX promotes progression of glioma through regulating miR-378e/RPN2 axis. J Exp Clin Cancer Res. 2019;38:506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 141. | Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu C, Li G, Zhu Y. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/β-catenin signaling pathway. Oncotarget. 2016;7:63449-63455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 177] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/