Published online Dec 20, 2023. doi: 10.5496/wjmg.v11.i4.39
Peer-review started: June 28, 2023
First decision: August 31, 2023
Revised: November 6, 2023
Accepted: November 30, 2023
Article in press: November 30, 2023
Published online: December 20, 2023
Processing time: 175 Days and 7.1 Hours
Pharmacogenomics (PG) testing is under-utilised in Australia. Our research provides Australia-specific data on the perspectives of patients who have had PG testing and those of the clinicians involved in their care, with the aim to inform wider adoption of PG into routine clinical practice.
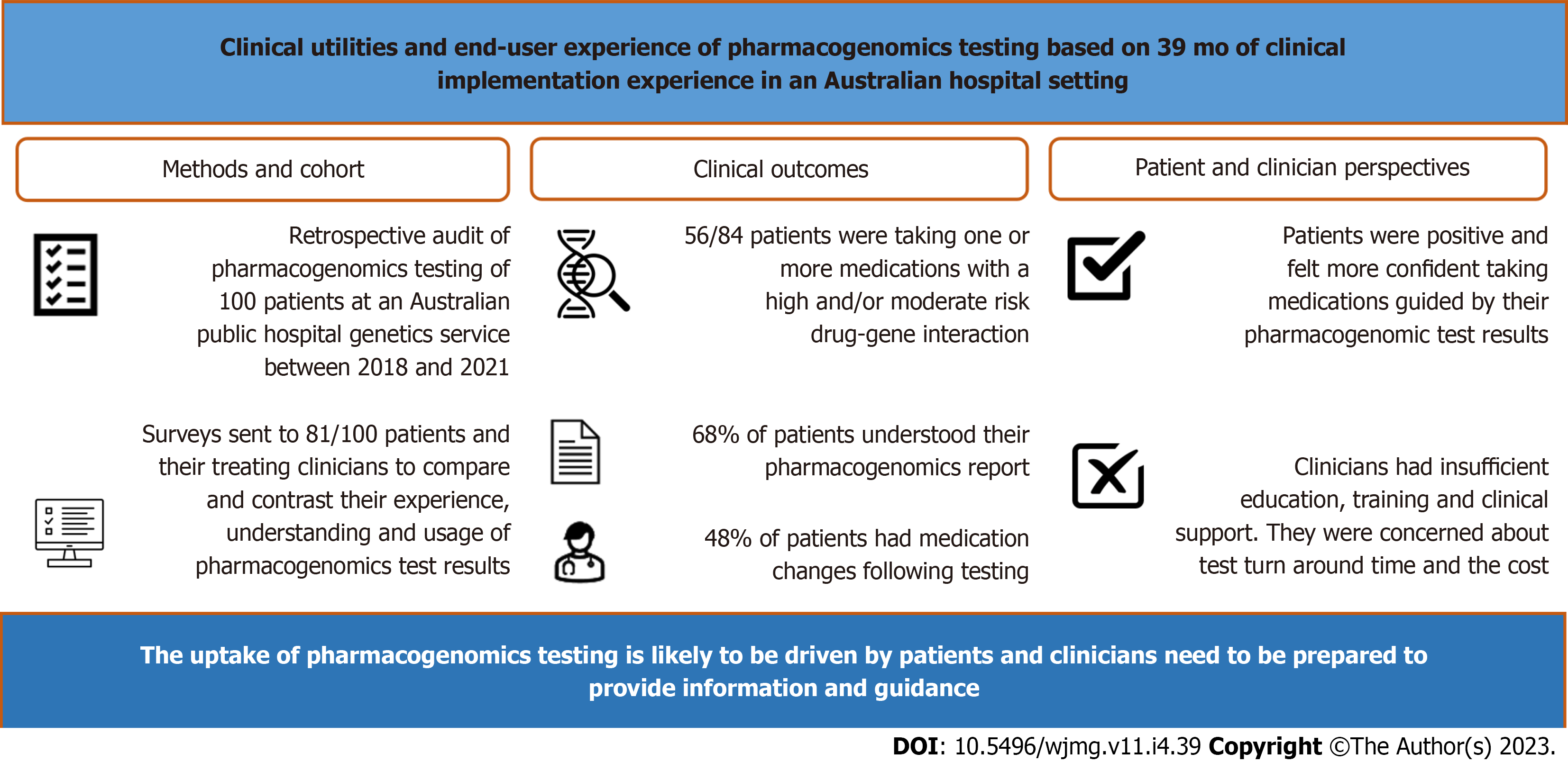
To investigate the frequency of actionable drug gene interactions and assess the perceived utility of PG among patients and clinicians.
We conducted a retrospective audit of PG undertaken by 100 patients at an Australian public hospital genetics service from 2018 to 2021. Via electronic surveys we compared and contrasted the experience, understanding and usage of results between these patients and their clinicians.
Of 100 patients who had PG, 84% were taking prescription medications, of which 67% were taking medications with actionable drug-gene interactions. Twenty-five out of 81 invited patients and 17 out of 89 invited clinicians completed the surveys. Sixty-eight percent of patients understood their PG results and 48% had medications changed following testing. Paired patient-clinician surveys showed patient-perceived utility and experience was positive, contrasting their clinicians’ hesitancy on PG adoption who identified insufficient education/training, lack of clinical support, test turnaround time and cost as barriers to adoption.
Our dichotomous findings between the perspectives of our patient and clinician cohorts suggest the uptake of PG is likely to be driven by patients and clinicians need to be prepared to provide information and guidance to their patients.
Core Tip: Pharmacogenomics (PG)-guided therapy has the potential to reduce adverse drug reactions and improve clinical outcomes of patients with mental illness. Despite increasing evidence, the uptake of PG among Australian clinicians remains low. We report for the first time on the dichotomous responses between patients and their clinician counterparts in the usage of PG: Patients were generally positive and willing to use PG compared with clinicians, suggesting that the uptake of PG in Australia is likely to be driven by patients, and that clinicians need to be prepared to provide information and guidance to their patients.
- Citation: Moxham R, Tjokrowidjaja A, Devery S, Smyth R, McLean A, Roberts DM, Wu KHC. Clinical utilities and end-user experience of pharmacogenomics: 39 mo of clinical implementation experience in an Australian hospital setting. World J Med Genet 2023; 11(4): 39-50
- URL: https://www.wjgnet.com/2220-3184/full/v11/i4/39.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v11.i4.39
Variabilities in an individual’s genetic sequence may affect the pharmacokinetics and pharmacodynamics of a drug, leading to an altered therapeutic response and/or increased risk of adverse drug reactions (ADRs)[1]. Pharmacogenomics (PG) is the study of clinically relevant genetic biomarkers, or pharmacogenetic variants that can influence a person’s response to therapeutic drugs[1]. The relationship between pharmacogenetic variants and drug response is termed drug-gene interaction (DGI). Recent advances in genomic technologies including Next Generation Sequencing, or Massively Parallel Sequencing, have led to an unprecedented capacity allowing multiple genes to be interrogated at once. This approach allows multiple genes that are involved in, not only different metabolising enzymes (pharmacokinetics) and drug receptors (pharmacodynamics) for a single drug, but also those of multiple drugs, to be analysed in a single test with a gene panel. Such pharmacogenomic testing can determine an individual’s likely response to multiple, up to a few hundred, medications from a single sample, unlike the traditional pharmacogenetic test of a single variant and single DGI at a time[2].
PG, especially if done pre-emptively, has the potential to guide clinicians at point-of-care in choosing the right therapeutic drug and dosing that is tailored to each patient, with the potential promises of reduced adverse effects, improved tolerability, and enhanced therapeutic response[3].
Internationally, there is a growing interest for widespread adoption of PG among patients and its potential to improve their medication outcomes[4-6]. In contrast, studies on clinician perspectives of PG have highlighted barriers and challenges towards its implementation into clinical practice. These barriers include lack of education/training, confidence, and acceptability of PG by clinicians[7-10], as well as a lack of consensus between clinical guidelines and access to clinical decision support[11-14]. To date, there is limited literature on the perspectives of the Australian end-users on PG[15].
In Australia, only two genotypes are covered by the national Medicare scheme, namely, HLA-B 5701 for abacavir, and TPMT for thiopurine drugs. However, multigene panel PG is neither publicly-funded by Medicare nor covered by private health insurance schemes. Patient-pay testing can be requested by a specialist or a general practitioner (GP). Most clinical genetics services in Australia, which are funded by State-operated public hospitals, do not routinely offer multigene panel PG, with the exception of St Vincent’s Hospital Sydney which is the setting of our investigation.
In Australia, the New South Wales Health Commission conducted an ‘Inquiry into the Management of Mental Heal
In this study we aim to: Investigate the potential impact of PG by determining the frequency of actionable DGIs in our patient cohort; assess within the same patient cohort, their experience of testing, as well as their understanding and usage of PG results and; assess among clinicians involved in the care of these patients, the experience, acceptability, under
A retrospective review was conducted on the PG reports and medication history of consecutive adult patients who were aged 18 years or older who had PG at St Vincent’s Hospital Clinical Genomics Sydney Australia between 1 August 2018 and 31 September 2021.
PG testing was conducted using a comprehensive commercial gene panel at a CLIA-certified laboratory (OneOme, Minneapolis, MN, United States), and included the following genes: CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, CYP4F2, COMP, DPYD, DRD2, F2, F5, GRIK4, HLA-A, HLA-B, HTR2A, HTR2C, IFNL4, NUDT15, OPRM1, SLC6A4, SLCO1B1, TPMT, UGT1A1, and VKORC1. This gene panel includes most pharmacogenes with high association evidence according to Clinical Pharmacogenetics Implementation Consortium, Dutch Pharmacogenetics Working Group and PG Knowledgebase[18].
Data collected on each patient included: age, sex, ethnicity, referring clinician and subspecialty, reason for referral, medication history, history of drug allergies and ADRs, number of high-risk and moderate-risk DGIs as shown on their PG report, number of current medications with high-risk or moderate-risk DGI.
Consecutive patients aged 18 years or older who underwent PG testing during a 39-month period between 1 August 2018 and 31 September 2021, and clinicians involved in the care of these patients, were identified and re-contacted via email and invited to participate in an online survey.
Patient and clinician surveys were developed and created on the Research Electronic Data Capture (REDCap) software. The patient survey assessed patient experience including their understanding and usage of the PG results. The clinician survey assessed clinician experience including their knowledge, acceptability and perceived utility of PG, as well as barriers to and support needs for broader adoption of PG in clinical practice. Demographic data were collected from both surveys. The surveys were constructed using 5-Point Likert Agreement Scale for categorical response questions, as well as other single or multiple option and open-ended comment questions. Survey questions were constructed using simple sentences at a literacy level of 5th grade using the Flesch-Kincaid Readability Calculator[19].
Both surveys were pilot tested by a focus group of five laypeople randomly identified from investigator’s peers and five clinicians who were randomly identified through St Vincent’s Hospital network, who were representative of our intended survey respondents. Members of the focus groups were provided with written instructions on how to review and test the survey and their feedback was sought regarding clarity, comprehension, functionality of the branching logic and the duration of time taken to complete the survey. Adjustments were made to the surveys with the final versions taking 15-20 min to complete. The REDCap surveys were distributed electronically via email, or short message service (SMS), if emails were unavailable, to patients and clinicians respectively. Follow-up reminder emails or SMSs were sent two weeks after the initial invitation if no responses were received.
Descriptive statistics were used to summarise categorical data ascertained from the retrospective chart review. If applicable, correlations were made between patient’s reported medication side effects/ADR and the DGI of the implicated medication. Mean, median, and standard deviation were derived using Microsoft Excel (Microsoft Corporation, 2016, version 16.0) for continuous variables such as age and number of high/moderate risk DGIs. The survey data were exported from REDCap and analysed using a regular analysis Microsoft Excel (Microsoft Corporation, 2016, version 16.0) package. Categorical data were summarised in descriptive and numerical formats using bar graphs and tables. For the 5-Point Likert scale (e.g. strongly agree, agree, neither agree nor disagree, disagree, strongly disagree), data was reviewed in its original form and combined into three categories (i.e. agree, neutral, disagree) then analysed as categorical data.
Ethics approval was sought and received from St Vincent’s Hospital Human Research Ethics Committee (2019
There were 100 patients identified during the audit period, consisting of 44 males and 56 females, with a mean age of 44.7 years +/- standard deviation 15.7 years (age range 18-75 years). Patients were referred by psychiatrists (39%), immunologists (15%), GPs (10%), neurologists (9%), transplant physicians (8%), and clinical pharmacologists (5%). The remaining 14% were referred by the following specialists: cardiologist, endocrinologist, gastroenterologist, haematologist, nephrologist, obstetrician, rheumatologist and thoracic physician; with each specialist category accounting for 2% of referrals. The referral indications were: to guide current pharmacotherapy (62%), previous history of ADRs (51%), history of polypharmacy (5%), and/or pre-emptive purpose to guide future prescription (5%). The demographics of the 100 patients are summarised in Table 1.
| Patient characteristics | |
| Sex | (%) |
| Male | 44 |
| Female | 56 |
| Age (yr) | (%) |
| 18-29 | 23 |
| 30-39 | 12 |
| 40-49 | 29 |
| 50-59 | 17 |
| 60-69 | 14 |
| > 70 | 5 |
| Referring specialists | (%) |
| Psychiatrists | 39 |
| Immunologists | 15 |
| General practitioners | 10 |
| Neurologists | 9 |
| Transplant physicians | 8 |
| Clinical pharmacologists | 5 |
| Others1 | 14 |
| Indications for referral | (%) |
| To guide pharmacotherapy | 62 |
| Previous drug reactions | 51 |
| Polypharmacy2 | 5 |
| Pre-emptive testing | 5 |
| Did not specify | 5 |
The PG reports of the 100 patients showed that on average, patients had 11 high-risk DGIs (range: 0-49, SD: 13.3) and 55 moderate-risk DGIs (range: 1-149, SD: 28.1). Of the 84 patients who provided a medication history, 17 patients (20%) were taking medications implicated with high-risk +/- moderate-risk DGI, 39 patients (46%) were taking medications with a moderate-risk DGI; with 56 patients (66%) taking medications with an actionable DGI (either high-risk DGI or moderate-risk DGI). Of these, one patient was taking nine medications with an actionable DGI (Table 2).
| Number of medications with implicated DGI | Number of patients taking medications with high-risk +/- moderate-risk DGI implicated | Number of patients taking medications with moderate-risk DGI implicated only | Number of patients taking medications with an actionable1 DGI |
| 1 | 5 (6) | 20 (24) | 25 (30) |
| 2 | 7 (8) | 11 (13) | 18 (21) |
| 3 | 3 (4) | 7 (8) | 10 (12) |
| 4 | 1 (1) | 0 (0) | 1 (1) |
| 5 | 0 (0) | 1 (1) | 1 (1) |
| 6 | 0 (0) | 0 (0) | 0 (0) |
| 7 | 0 (0) | 0 (0) | 0 (0) |
| 8 | 0 (0) | 0 (0) | 0 (0) |
| 9 | 1 (1) | 0 (0) | 1 (1) |
| 10 | 0 (0) | 0 (0) | 0 (0) |
| Total | 17 (20) | 39 (46) | 56 (66) |
Of the 100 patients identified and included in the retrospective review, 81 patients were invited to participate in the patient survey, after excluding those with no contact details and those who had indicated they did not wish to be contacted for future research. Of the 81 patients invited, 25 patients (31%) responded and completed the survey. Patient survey respondents were mostly female, aged 40-49 years with a tertiary level of education. A total of 89 clinicians, including 29 subspecialist clinicians and 60 GPs, involved in the care of the same patient cohort were identified and invited to participate in the clinician survey. Of those invited, 17 clinicians (19%) responded and completed the survey. Clinician survey respondents were mostly male, practicing in psychiatry, with a medical degree attained after the year 2000, who encountered patients in the hospital setting and had referred or discussed PG with their patients in the preceding 12 mo. A summary of survey respondent demographics for both patient and clinician surveys is shown in Table 3.
| Patient respondents (n = 25) | Clinician respondents (n = 17) | ||
| Sex | Sex | ||
| Male | 9 (36.0) | Male | 11 (64.7) |
| Female | 16 (64.0) | Female | 6 (35.3) |
| Patient age (yr) | Clinician attained medical degree | ||
| 18-29 | 4 (16.0) | Before 1980 | 2 (11.8) |
| 30-39 | 2 (8.0) | 1981-1990 | 2 (11.8) |
| 40-49 | 7 (28.0) | 1991-2000 | 3 (17.6) |
| 50-59 | 6 (24.0) | After 2000 | 10 (58.8) |
| 60-69 | 5 (20.0) | ||
| > 70 | 1 (4.0) | ||
| Patient education level | Clinician specialty | ||
| Did not finish high school | 1 (4.0) | Psychiatry | 5 (29.4) |
| High school | 6 (24.0) | Clinical pharmacology | 3 (17.6) |
| Certificate or diploma | 5 (20.0) | Immunology | 2 (11.8) |
| Bachelor’s degree | 11 (44.0) | Neurology | 2 (11.8) |
| Master’s degree or doctorate | 2 (8.0) | General practitioner | 1 (5.9) |
| Other1 | 7 (41.2) |
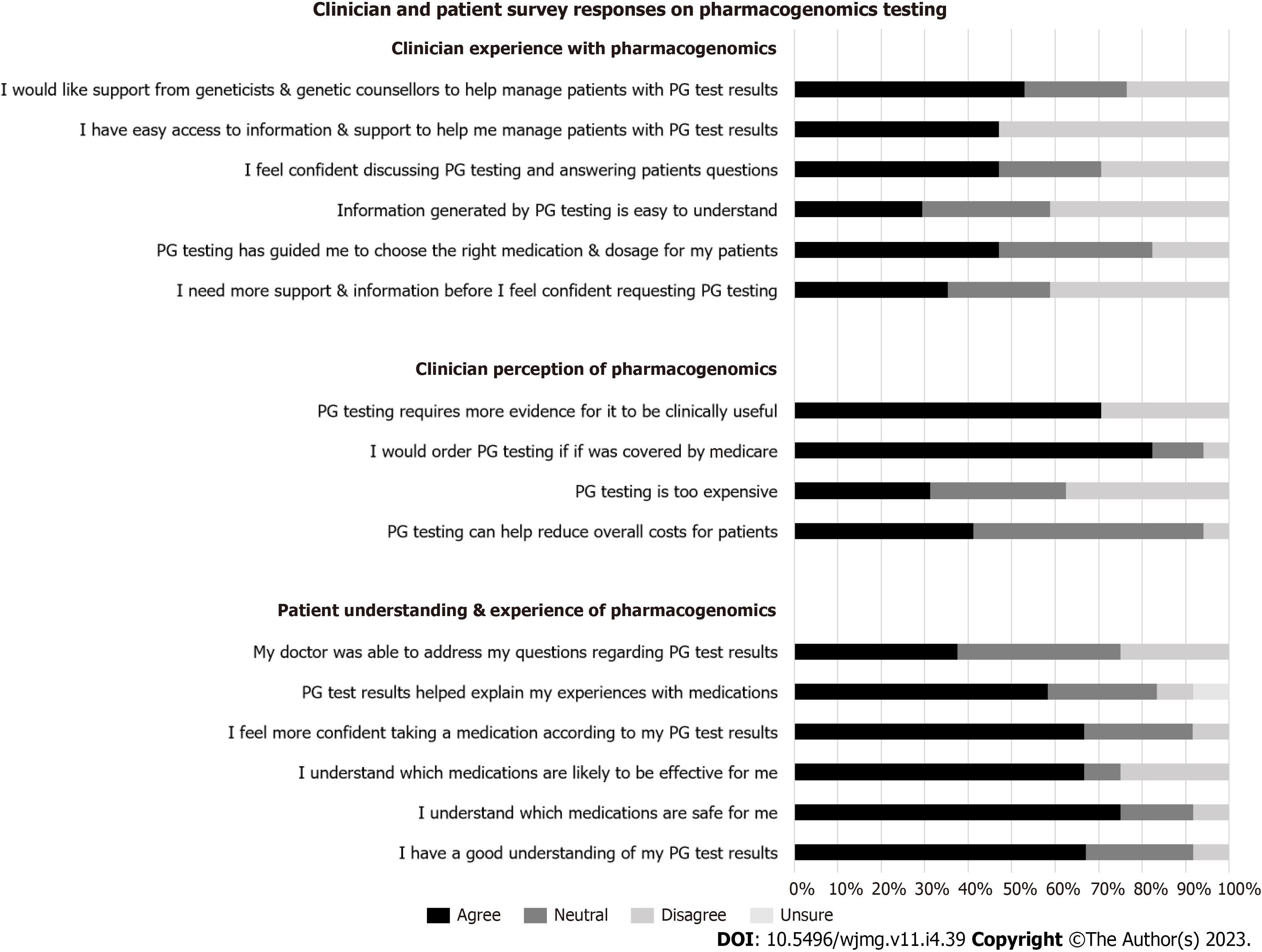
Responses from clinician survey (n = 17) showed that clinicians have different levels of knowledge and sources of education for PG. The majority of clinicians (94.1%) knew that individuals respond differently to medications based on genetic factors. Their knowledge of how many drugs have a PG impact varied significantly. None of the clinicians had completed any formal training in PG, with the majority (52.9%) reporting not feeling confident in discussing and/or answering questions about PG with their patients. The primary sources of knowledge about PG for clinicians were medical colleagues (75%), scientific journals (62.5%), conferences and scientific meetings (25%), publicly available online resources (25%) and/or commercial providers such as pathology laboratories (18.8%).
Patient experience: Patient respondents (n = 25) were generally positive in their experience with PG. The majority of patients (68%) agreed with the statement “I have a good understanding of my PG results”. Nineteen (76%) and 17 (68%) patient respondents agreed that PG has helped them understand which medications were safe, and effective, for them respectively. Likewise, 17 patients (68%) agreed with the statement “I feel more confident in a new medication chosen by my doctor according to my PG results”; and 15 patients (60%) agreed with the statement “The results of my PG test results helped explain my experiences with medications”. Nine patients (36%) agreed with the statement “My doctor was able to address my questions regarding my PG results” (Figure 2).
Clinician perspective: Clinician respondents (n = 17) were generally ambivalent in their perception of PG utility, with 50% reporting that PG has benefitted some of their patients, while 37.5% and 6.3% reported that PG has benefitted little, or none, of their patients, respectively; while 6.3% reported it has benefitted most of their patients. In addition, 17.7% of respondents disagreed with, and 35.3% were neutral to, the statement “PG has guided me in choosing the right medication and/or dosage for my patients”. When asked about the likelihood of utilising PG in the next 12 mo, 64.7% clinicians reported likely, 23.5% reported unlikely, and 11.8% were ambivalent about using it in the next 12 mo. Seven (41.2%) clinicians agreed that “PG can help reduce overall costs for patients”, while nine (52.9%) were neutral to the same statement. There was an equal distribution among clinicians who agreed with (31.3%), were neutral to (31.3%), or disagreed with (37.6%), the statement that PG was too expensive for their patients. Most clinicians (88.2%) agreed that they were more likely to order PG testing if Medicare covered the cost (Figure 2).
As shown in Figure 3, the majority of patient respondents (92%) have shared their PG results with their health care providers; and 12 patients (48%) reported having had their medications changed over 17 clinical encounters following PG testing. Of the seventeen medication changes reported by patient respondents, nine instances (53%) of medication changes were made by their specialist, seven (41%) by their GP, and one instance (6%) was made by the patient themselves.
As shown in Figure 2, clinicians were ambivalent towards the adoption of PG. The majority of clinician respondents (70.6%) thought that PG requires more evidence for it to be clinically useful. The majority of respondents (70.6%) did not agree that the information generated from PG was easy to understand; the majority (52.9%) did not feel confident in discussing and answering questions about PG. When asked if support is needed before clinicians feel comfortable in requesting PG, the responses were fairly equally distributed, between agree (35.3%), neutral (23.5%), and disagree (41.2%). The majority of clinician respondents (52.9%) reported not having easy access to resources to help manage their patients; and 52.9% respondents would like support from genetic professionals to help manage patients who have undergone PG testing.
In regards to whom clinicians thought would be the most appropriate person to discuss and return PG results to patients, the responses were equally divided between: Specialist ordering the test (52.9%), and clinical geneticists/genetic counsellors (47.1%); with no responses for GPs or pharmacists.
Barriers to adoption of PG were identified from our clinician cohort whose patients have had PG testing. The single most important barrier as reported by clinician respondents includes one of: A lack of clinical decision aids (29.4%), cost factor for patients (23.5%), timeliness of results (23.5%), lack of access to information (11.8%), patient expectations (5.9%) and difficult logistics in organising testing (5.9%). If barriers were overcome, most but not all clinicians (76.5%) agreed there was value in PG for their patients, with 17.6% providing a neutral response and 5.9% disagreeing to the statement.
Despite recent advances in genomic testing, the increasing knowledge of DGIs, and the availability of dosing guidelines based on actionable DGIs[20], PG remains under-utilised in Australia. There have been several PG implementation projects undertaken in the US and Europe; such initiatives are limited in Australia[21].
Our study highlighted the potential utility of PG in identifying high- and/or moderate-risk, or actionable, DGIs that may trigger a change in prescription. Our audit showed a wide range of variabilities in the number and degree of DGIs among patients, supporting the rationale for individualised prescribing based on genetic characteristics. Additionally, our results indicated that a significant proportion of patients, 66%, were taking a medication implicated to have an actionable DGI at the time of PG testing, highlighting the potential utility of PG in guiding prescription and reducing the incidence of adverse drug effects[22].
Although our patients who have had testing were generally in favour of the utility of PG on improving their medication experience, the perception of PG among clinicians who care for these patients was however reserved with hesitancy and/or skepticism. Over half of our patient respondents (68%) reported feeling more confident in medications when their PG results were taken into consideration by their healthcare providers when prescribing. This echoes the limited literature showing that PG may increase medication adherence among patients[23]. In contrast to patient per
Half of the patient respondents in our study reported having their medications changed following PG, highlighting the potential impact of PG on treatment decisions. Additionally, our patient survey showed that the majority of respondents have discussed their PG results with their GPs; and of those who had medication changes, half of the respondents had their medications changed by their GPs. These findings suggest the importance of primary care engagement and involvement in the wider adoption and implementation of PG in Australia.
We identified a lack of education and training, and a lack of clinical decision aids and support as the major barriers to routine adoption of PG among our clinician cohort who was involved in the care of patients who underwent PG testing. The majority of our clinicians have not completed any formal training in PG; have expressed complexities of incor
Other barriers to uptake of PG identified in our study include time limitations and cost of testing. The long turnaround time for results highlights the importance of testing in a pre-emptive setting so that results are available at future point-of-care. In Australia, Medicare currently covers only two DGIs (HLA-B 5701 for abacavir, TPMT for thiopurine drugs). Most clinicians would like to see more PG tests covered by Medicare and reported that they are more likely to order a test if there was public funding. From the patient perspective, our patient survey did not capture cost implications as testing was undertaken at no charge to patients. To date, economic studies have suggested that PG testing can be cost-effective and that the cost of testing could be offset by its cost-savings from reduced time wastage on medication trial-and-error, enhanced therapeutic response, and mitigation of ADRs[30-32]. Future studies that explore the cost implications of PG-informed prescription should capture data on, not only individual patients, but also the overall healthcare system, to inform public funding for mainstream implementation in Australia.
The present study is limited by a lack of generalizability of our survey results, due to the small sample size, with 72% of patient respondents having a tertiary qualification or higher, and recruitment bias through a single tertiary genetics service. Similarly, our clinician respondents represent a small niche cohort who mostly work in a tertiary hospital setting. Our survey response rate of 31% from patients and 19% from clinicians in a sample size smaller than 500 can be considered sufficient to conclude estimates but is still below the average online survey response rate of 44.1% and is therefore best considered as observatory and exploratory[33]. Furthermore, our study had limited, 5.9%, survey response rate from GPs, which may be attributable to the coronavirus disease 2019 pandemic impact during the time of the survey. This makes it difficult to draw perspectives from this clinician cohort who is potentially a frontline user and adopter of PG. As a Cronbach’s Alpha Test was not performed to assess the reliability or internal consistency of the survey questions these surveys were not considered validated[34]. Finally, recall bias is an inherent limitation of survey research into past experiences, in particular, patient recall of medication changes, as well as understanding and sharing of PG results.
This study provides not only Australia-specific data, but also the perspectives of patients who have had PG and those of the clinicians involved in their care, which can inform future research into the implementation and the wider adoption of PG in Australia. Through an audit of 100 patients who have had testing, we found a significant proportion, 66%, of patients were taking a medication with an actionable DGI. Our paired end-user surveys highlighted dichotomous perspectives on PG between patients and their clinicians. Patients were generally positive and willing to use PG compared with clinicians, suggesting that patients may play a significant role in driving change and promoting the uptake of PG in clinical care.
In Australia, the New South Wales Health Commission conducted an ‘Inquiry into the Management of Mental Healthcare Delivery’ in 2018 that identified under-utilisation of pharmacogenomics (PG) and recommended such testing as a key priority to improve clinical outcomes for mental health patients. In order to see wider uptake, one needs to identify barriers to and facilitators of PG adoption in Australia, as perceived by patients who have had testing and clinicians who care for these patients. Such data, which remain limited to date, will inform an implementation science approach to promote the systematic uptake of PG into routine practice.
PG-guided therapy has been shown to significantly enhance clinical outcomes in the field of mental health. Currently, two-thirds of patients with a major depressive disorder fail to achieve remission during the first treatment level; and many require multiple different medications sequentially on a trial-and-error basis. In addition, it has been shown that the odds of remission diminish with every additional medication trial-and-error iteration; and that a window of therapeutic opportunity for major depressive disorder appears to be within the first 2 sequential treatments. PG-guided pharmacotherapy, which aims at giving the right drug at the right dosage to the right person at the right intervention time, has the potential to enhance both patient outcomes and health cost savings in the Australian mental health system. Despite the evidence, Australia has been slow in adopting PG testing to guide therapy. To facilitate widespread clinical implementation of PG it is necessary to know and understand the attitudes and acceptability of PG, as well as the support and resource requirements, amongst potential end-users, including clinicians and patients. This information can contribute to the development of a model of care, enhance the feasibility and clinical utility of PG to ensure successful implementation of PG-guided pharmacotherapy into routine clinical care of patients with mental illness.
The objectives were to evaluate retrospective file data of patients participating in PG testing in Australia and to re-contact patients and their general practitioners (GPs)/clinicians to assess the impact of PG both on treatment options and patient/clinician reported outcomes.
A retrospective audit was undertaken of the PG results of 100 patients who underwent testing as part of their routine clinical care between 1 August 2018 and 31 September 2021 at an Australian public hospital genetics service. Specifically designed and pilot-tested surveys were used to assess clinician knowledge, acceptability and perceived utility of PG in the care of patients and evaluate patients knowledge of PG results, as well as, how the PG has impacted their pharmaceutical care and overall mental and physical health. Data was analysed using descriptive statistics to summarise categorical data ascertained from the retrospective chart review where correlations were made between patient’s reported medication side effects/adverse drug reaction (ADR) and the drug-gene interaction (DGI) of the implicated medication. Categorical data from clinician and patient surveys were summarised in descriptive and numerical formats with bar graphs and tables.
Through an audit of 100 patients who have had PG testing at St Vincent’s Hospital Clinical Genomics Sydney, we found 67% of patients were taking a medication with an actionable DGI at the time of testing. The importance of our work is that via paired end-user surveys it highlighted dichotomous perspectives on PG between patients and their clinicians (psychiatrists, immunologists, neurologists, transplant physicians, clinical pharmacologists and GPs). Patients were generally in favour of the utility of PG on improving their medication experience, the perception of PG among clinicians who care for these patients was however reserved with hesitancy and/or skepticism. This suggests that the uptake of PG is likely to be driven by patients, and clinicians need to be prepared to provide information and guidance to their patients.
Our research identifies barriers to the clinical implementation of PG and suggests strategic solutions that could be put in place to support a wider adoption in routine medical practice. The majority (70%) of our patient participants have discussed their PG results with their GP; and of those who had medication changes following testing, half of the respondents had their medications changed by their GP. These findings suggest the importance of primary care involvement in the wider adoption and implementation of PG in Australia.
To date, economic studies have suggested that PG testing can be cost-effective and that the cost of testing could be offset by its cost-savings from reduced time wastage on medication trial-and-error iterations, enhanced therapeutic response, and mitigation of ADRs. Future studies that explore the cost implications of PG-informed prescription should capture data on, not only individual patients, but also the overall healthcare system, to inform public funding for mainstream implementation in Australia.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: Human Genetics Society of Australasia (HGSA); Royal Australasian College of Physicians (RACP); St Vincent’s Hospital Clinical Genomics, Sydney, NSW, Australia; School of Clinical Medicine, Faculty of Medicine and Health, University of New South Wales Sydney, Australia; School of Medicine, University of Notre Dame Australia, Sydney, NSW, Australia; Discipline of Genetic Medicine, University of Sydney, Sydney, NSW, Australia. Past member of American Society of Human Genetics (ASHG).
Specialty type: Genetics and heredity
Country/Territory of origin: Australia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Belančić A, Croatia S-Editor: Qu XL L-Editor: A P-Editor: Xu ZH
| 1. | Kozyra M, Ingelman-Sundberg M, Lauschke VM. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet Med. 2017;19:20-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 167] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 2. | Nebert DW, Zhang G, Vesell ES. From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab Rev. 2008;40:187-224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 3. | Haidar CE, Crews KR, Hoffman JM, Relling MV, Caudle KE. Advancing Pharmacogenomics from Single-Gene to Preemptive Testing. Annu Rev Genomics Hum Genet. 2022;23:449-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 4. | Rogausch A, Prause D, Schallenberg A, Brockmöller J, Himmel W. Patients' and physicians' perspectives on pharmacogenetic testing. Pharmacogenomics. 2006;7:49-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 93] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 5. | Haddy CA, Ward HM, Angley MT, McKinnon RA. Consumers' views of pharmacogenetics--A qualitative study. Res Social Adm Pharm. 2010;6:221-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 6. | Olson JE, Rohrer Vitek CR, Bell EJ, McGree ME, Jacobson DJ, St Sauver JL, Caraballo PJ, Griffin JM, Roger VL, Bielinski SJ. Participant-perceived understanding and perspectives on pharmacogenomics: the Mayo Clinic RIGHT protocol (Right Drug, Right Dose, Right Time). Genet Med. 2017;19:819-825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 7. | St Sauver JL, Bielinski SJ, Olson JE, Bell EJ, Mc Gree ME, Jacobson DJ, McCormick JB, Caraballo PJ, Takahashi PY, Roger VL, Rohrer Vitek CR. Integrating Pharmacogenomics into Clinical Practice: Promise vs Reality. Am J Med. 2016;129:1093-1099.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 8. | Söyletir G, Babacan F, Göral M, Can A. [Prevalence of anti-delta antibodies in HBsAg carriers]. Mikrobiyol Bul. 1989;23:97-101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 9. | Green JS, O'Brien TJ, Chiappinelli VA, Harralson AF. Pharmacogenomics instruction in US and Canadian medical schools: implications for personalized medicine. Pharmacogenomics. 2010;11:1331-1340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Rohrer Vitek CR, Giri J, Caraballo PJ, Curry TB, Nicholson WT. Pharmacogenomics education and perceptions: is there a gap between internal medicine resident and attending physicians? Pharmacogenomics. 2021;22:195-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 11. | Nicholson WT, Formea CM, Matey ET, Wright JA, Giri J, Moyer AM. Considerations When Applying Pharmacogenomics to Your Practice. Mayo Clin Proc. 2021;96:218-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 12. | Amara N, Blouin-Bougie J, Bouthillier D, Simard J. On the readiness of physicians for pharmacogenomics testing: an empirical assessment. Pharmacogenomics J. 2018;18:308-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 13. | Shugg T, Pasternak AL, London B, Luzum JA. Prevalence and types of inconsistencies in clinical pharmacogenetic recommendations among major U.S. sources. NPJ Genom Med. 2020;5:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 14. | Guo C, Hu B, Guo C, Meng X, Kuang Y, Huang L, Wang D, Xu K, Zhao Y, Yang G, Cai W, Shu Y. A Survey of Pharmacogenomics Testing Among Physicians, Pharmacists, and Researchers From China. Front Pharmacol. 2021;12:682020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Suppiah V, Lim CX, Hotham E. Community pharmacists and their role in pharmacogenomics testing: an Australian perspective drawing on international evidence. Aust J Prim Health. 2018;24:441-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Public Accounts Committee 2018. Inquiry into the management of health care delivery in NSW. [cited 19 March 2021]. Available from: https://www.parliament.nsw.gov.au/committees/inquiries/Pages/inquiry-details.aspx?pk=2423. |
| 17. | O'Shea R, Ma AS, Jamieson RV, Rankin NM. Precision medicine in Australia: now is the time to get it right. Med J Aust. 2022;217:559-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 18. | Alshabeeb MA, Alyabsi M, Aziz MA, Abohelaika S. Pharmacogenes that demonstrate high association evidence according to CPIC, DPWG, and PharmGKB. Front Med (Lausanne). 2022;9:1001876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Flesch-Kincaid Readability Calculator. [cited 20 July 2022]. Available from: https://goodcalculators.com/flesch-kincaid-calculator/. |
| 20. | US Department of Health and Human Services. Clinical Pharmacogenetics Implementation Consortium Guidelines. [cited 7 November 2022]. Available from: https://cpicpgx.org/guidelines/. |
| 21. | Krebs K, Milani L. Translating pharmacogenomics into clinical decisions: do not let the perfect be the enemy of the good. Hum Genomics. 2019;13:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 121] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 22. | White C, Scott R, Paul CL, Ackland SP. Pharmacogenomics in the era of personalised medicine. Med J Aust. 2022;217:510-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 23. | Christian C, Borden BA, Danahey K, Yeo KJ, van Wijk XMR, Ratain MJ, O'Donnell PH. Pharmacogenomic-Based Decision Support to Predict Adherence to Medications. Clin Pharmacol Ther. 2020;108:368-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Unertl KM, Jaffa H, Field JR, Price L, Peterson JF. Clinician Perspectives on Using Pharmacogenomics in Clinical Practice. Per Med. 2015;12:339-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 25. | Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians' knowledge of and experience with pharmacogenetic testing. Clin Genet. 2012;82:388-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 215] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 26. | Aquilante CL, Kao DP, Trinkley KE, Lin CT, Crooks KR, Hearst EC, Hess SJ, Kudron EL, Lee YM, Liko I, Lowery J, Mathias RA, Monte AA, Rafaels N, Rioth MJ, Roberts ER, Taylor MR, Williamson C, Barnes KC. Clinical implementation of pharmacogenomics via a health system-wide research biobank: the University of Colorado experience. Pharmacogenomics. 2020;21:375-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 27. | Dunnenberger HM, Biszewski M, Bell GC, Sereika A, May H, Johnson SG, Hulick PJ, Khandekar J. Implementation of a multidisciplinary pharmacogenomics clinic in a community health system. Am J Health Syst Pharm. 2016;73:1956-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 28. | Caraballo PJ, Hodge LS, Bielinski SJ, Stewart AK, Farrugia G, Schultz CG, Rohrer-Vitek CR, Olson JE, St Sauver JL, Roger VL, Parkulo MA, Kullo IJ, Nicholson WT, Elliott MA, Black JL, Weinshilboum RM. Multidisciplinary model to implement pharmacogenomics at the point of care. Genet Med. 2017;19:421-429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 77] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 29. | Pearce A, Terrill B, Alffenaar JW, Patanwala AE, Kummerfeld S, Day R, Young MA, Stocker SL. Pharmacogenomic testing: perception of clinical utility, enablers and barriers to adoption in Australian hospitals. Intern Med J. 2022;52:1135-1143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 30. | Verbelen M, Weale ME, Lewis CM. Cost-effectiveness of pharmacogenetic-guided treatment: are we there yet? Pharmacogenomics J. 2017;17:395-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 165] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 31. | Zhu Y, Swanson KM, Rojas RL, Wang Z, St Sauver JL, Visscher SL, Prokop LJ, Bielinski SJ, Wang L, Weinshilboum R, Borah BJ. Systematic review of the evidence on the cost-effectiveness of pharmacogenomics-guided treatment for cardiovascular diseases. Genet Med. 2020;22:475-486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 32. | Tanner JA, Davies PE, Overall CC, Grima D, Nam J, Dechairo BM. Cost-effectiveness of combinatorial pharmacogenomic testing for depression from the Canadian public payer perspective. Pharmacogenomics. 2020;21:521-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 33. | Wu M-J, Zhao K, Fils-Aime F. Response rates of online surveys in published research: A meta-analysis. Com in Hum Beh Rep. 2022;7:100206. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 411] [Article Influence: 102.8] [Reference Citation Analysis (0)] |
| 34. | Taber K. The use of Cronbach’s Alpha when developing and reporting research instruments in science education. Res Sci Educ. 2018;48:1273-1296. [DOI] [Full Text] |