©The Author(s) 2025.
World J Exp Med. Dec 20, 2025; 15(4): 113869
Published online Dec 20, 2025. doi: 10.5493/wjem.v15.i4.113869
Published online Dec 20, 2025. doi: 10.5493/wjem.v15.i4.113869
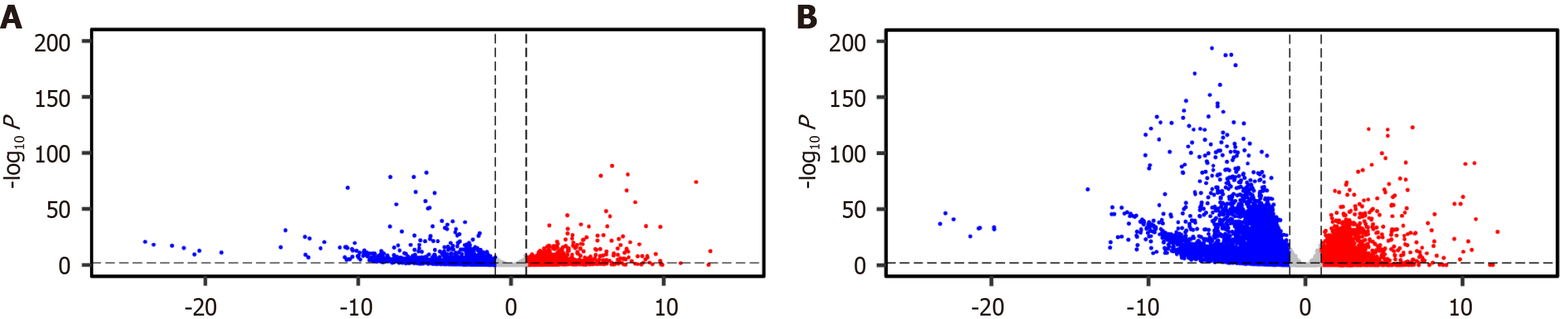
Figure 1 Transcriptome-wide differential expression profiles in vaccine-affected individuals.
A: Volcano plot showing differential gene expression in individuals with new-onset adverse events following messenger RNA (mRNA) coronavirus disease 2019 (COVID-19) vaccination (n = 3) compared to normal controls (n = 803); B: Volcano plot showing differential gene expression in individuals diagnosed with new-onset cancer shortly after receiving mRNA COVID-19 vaccination (n = 7) compared to the same control cohort. Each point represents a single gene plotted by log2 fold change (X-axis) and -log10 adjusted P value (Padj) (Y-axis). Genes with significant upregulation (log2FC > 1, Padj < 0.05) are marked in red, while significantly downregulated genes (log2FC < -1, Padj < 0.05) are shown in blue. Non-significant genes appear in gray. These plots reveal widespread transcriptional dysregulation in both patient groups, serving as the foundation for subsequent pathway enrichment analysis.
Figure 2 Protein-protein interaction network of the most dysregulated genes in group 1.
A: Genes related to proliferative signaling and tumor control; B: Genes related to systemic inflammatory immune; C: Genes related to transcriptomic instability; D: Genes related to mitochondrial electron transport dysfunction and reactive oxygen species; E: Genes related to proteasome-mediated protein degradation stress. Group 1 includes patients with new-onset adverse events following messenger RNA coronavirus disease 2019 vaccination (n = 3), compared with normal controls (n = 803 unvaccinated individuals from the GTEx dataset). Node color intensity reflects the degree of interaction (connectivity), with darker nodes indicating higher connectivity or a hub status within the network.
Figure 3 Protein-protein interaction network of the most dysregulated genes in group 2.
A: Genes related to transcriptomic instability, translational stress; B: Genes related to genomic instability and epigenetic shift; C: Genes associated with endothelial dysfunction; D: Genes related to systemic inflammatory and immune response; E: Genes that are related to proliferative signaling and suppressed tumor control. Group 2 includes patients with new-onset cancers diagnosed shortly after messenger RNA coronavirus disease 2019 vaccination (n = 7), compared with normal controls (n = 803 unvaccinated individuals from the GTEx dataset). Node color intensity reflects the degree of interaction (connectivity), with darker nodes indicating higher connectivity or a hub status within the network.
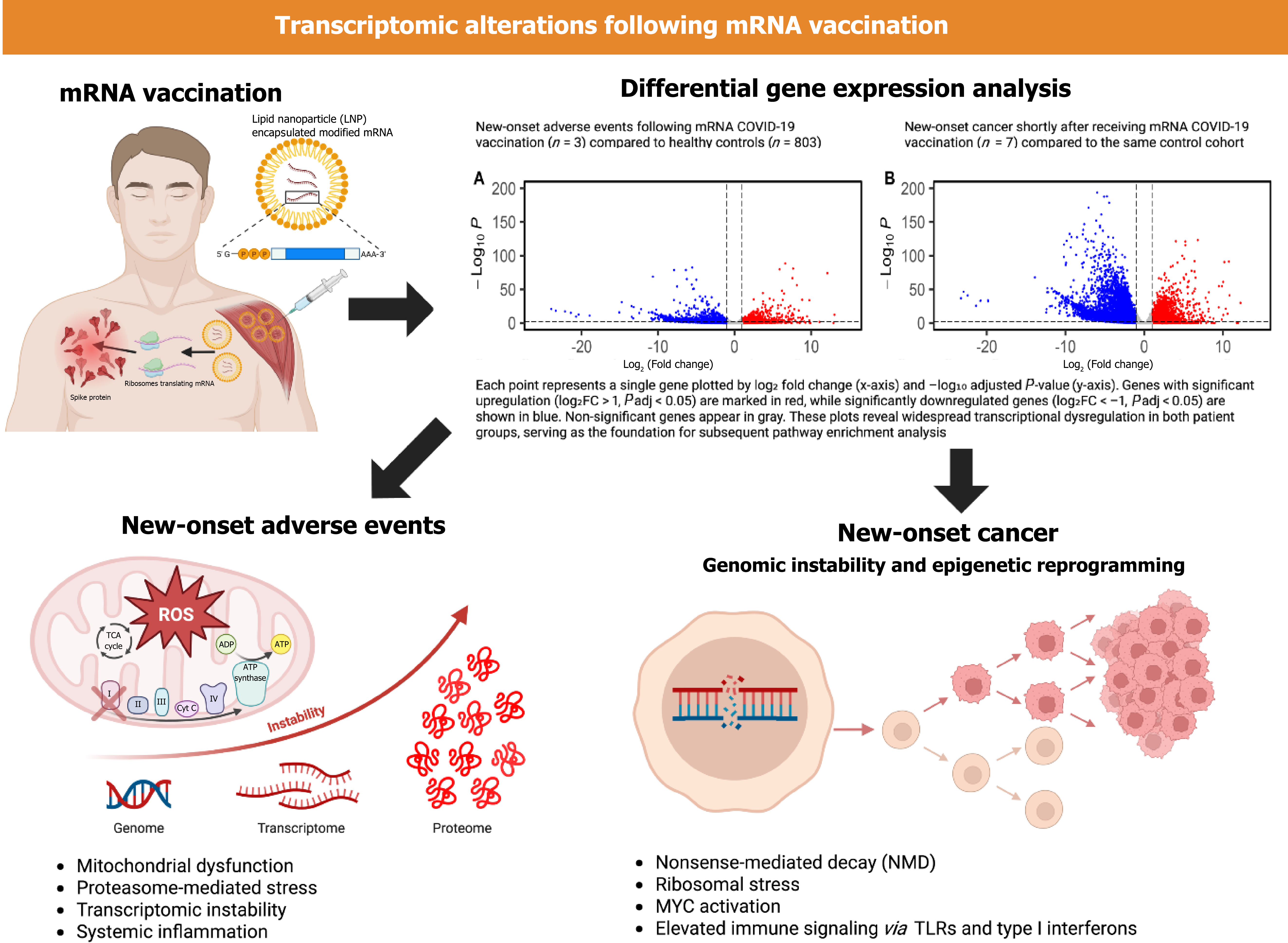
Figure 4 Transcriptomic alterations following messenger RNA vaccination.
This central illustration summarizes the experimental design, core findings, and proposed molecular mechanisms underlying transcriptomic dysregulation following synthetic messenger RNA (mRNA) coronavirus disease 2019 vaccination. Top left: Schematic of mRNA vaccination, showing lipid nanoparticle-encapsulated, chemically modified mRNA encoding spike protein delivered into host cells, initiating persistent translation and immunologic engagement. Top right: Volcano plots depict global differential gene expression in peripheral blood samples from two affected cohorts vs normal controls (n = 803). Left: Individuals with new-onset nonmalignant adverse events (n = 3). Right: Individuals with new-onset cancer (n = 7). Upregulated genes [log2FC > 1, adjusted P value (Padj) < 0.05] are shown in red; downregulated genes (log2FC < -1, Padj < 0.05) in blue; non-significant genes in gray. Bottom left (new-onset adverse events): Transcriptomic analysis reveals enrichment of pathways linked to mitochondrial electron transport dysfunction and reactive oxygen species, proteasome-mediated protein degradation stress, mRNA surveillance activation, and systemic inflammatory signaling. Bottom right (new-onset cancer): Cancer patients exhibit hallmarks of oncogenesis, including genomic instability, epigenetic reprogramming, nonsense-mediated decay, ribosomal stress, myelocytomatosis oncogene-driven proliferative signaling, and persistent immune activation via toll-like receptors and type I interferons. COVID-19: Coronavirus disease 2019; mRNA: Messenger RNA; MYC: Myelocytomatosis oncogene; ROS: Reactive oxygen species; TCA: Tricarboxylic acid; TLRs: Toll-like receptors;
- Citation: Von Ranke NL, Zhang W, Anokhin P, Hulscher N, McKernan K, Mccullough P, Catanzaro J. Synthetic messenger RNA vaccines and transcriptomic dysregulation: Evidence from new-onset adverse events and cancers post-vaccination. World J Exp Med 2025; 15(4): 113869
- URL: https://www.wjgnet.com/2220-315x/full/v15/i4/113869.htm
- DOI: https://dx.doi.org/10.5493/wjem.v15.i4.113869