Published online Aug 20, 2015. doi: 10.5493/wjem.v5.i3.164
Peer-review started: February 26, 2015
First decision: April 27, 2015
Revised: May 21, 2015
Accepted: June 9, 2015
Article in press: June 11, 2015
Published online: August 20, 2015
Processing time: 181 Days and 3.8 Hours
Tuberculosis is one of the leading infectious diseases plaguing mankind and is mediated by the facultative pathogen, Mycobacterium tuberculosis (MTB). Once the pathogen enters the body, it subverts the host immune defenses and thrives for extended periods of time within the host macrophages in the lung granulomas, a condition called latent tuberculosis (LTB). Persons with LTB are prone to reactivation of the disease when the body’s immunity is compromised. Currently there are no reliable and effective diagnosis and treatment options for LTB, which necessitates new research in this area. The mycobacterial proteins and genes mediating the adaptive responses inside the macrophage is largely yet to be determined. Recently, it has been shown that the mce operon genes are critical for host cell invasion by the mycobacterium and for establishing a persistent infection in both in vitro and in mouse models of tuberculosis. The YrbE and Mce proteins which are encoded by the MTB mce operons display high degrees of homology to the permeases and the surface binding protein of the ABC transports, respectively. Similarities in structure and cell surface location impute a role in cell invasion at cholesterol rich regions and immunomodulation. The mce4 operon is also thought to encode a cholesterol transport system that enables the mycobacterium to derive both energy and carbon from the host membrane lipids and possibly generating virulence mediating metabolites, thus enabling the bacteria in its long term survival within the granuloma. Various deletion mutation studies involving individual or whole mce operon genes have shown to be conferring varying degrees of attenuation of infectivity or at times hypervirulence to the host MTB, with the deletion of mce4A operon gene conferring the greatest degree of attenuation of virulence. Antisense technology using synthetic siRNAs has been used in knocking down genes in bacteria and over the years this has evolved into a powerful tool for elucidating the roles of various genes mediating infectivity and survival in mycobacteria. Molecular beacons are a newer class of antisense RNA tagged with a fluorophore/quencher pair and their use for in vivo detection and knockdown of mRNA is rapidly gaining popularity.
Core tip: This review paper looks at the current status of research of the role of mammalian cell entry gene products in mediating cholesterol mediated latency of mycobacteria and the potential use of short-interfering RNA molecular beacons in detecting and attenuating mycobacterial infections.
- Citation: George R, Cavalcante R, Jr CC, Marques E, Waugh JB, Unlap MT. Use of siRNA molecular beacons to detect and attenuate mycobacterial infection in macrophages. World J Exp Med 2015; 5(3): 164-181
- URL: https://www.wjgnet.com/2220-315X/full/v5/i3/164.htm
- DOI: https://dx.doi.org/10.5493/wjem.v5.i3.164
The year 2005 marked the 100th anniversary since Robert Koch received the Nobel prize for his work on tuberculosis (TB) and yet more than one hundred years later the World Health Organization (WHO) has reaffirmed its designation of TB as a global emergency[1]. Tuberculosis still remains a pandemic, infecting one-third of the world’s population and killing millions of people each year. Estimates are that a TB death occurs every minute. According to recent estimates of WHO, nearly 9 million people were infected with TB in 2012, including 1.3 million TB-related deaths worldwide[2]. Incidentally, TB and reactivation of latent TB have turned out to be the leading causes of death for people who are infected with human immunodeficiency virus (HIV). At the same time, according to the Centers for Disease Control (CDC), there were 9945 TB cases reported in the United States in 2012[3]. More than 80% of TB cases in the United States are from reactivation of latent TB infection[4].
Tuberculosis is a disease that spreads from person to person through the air and is mediated by the pathogen Mycobacterium tuberculosis (MTB). The TB bacillus was discovered in 1882 and has been the subject of extensive research since then. There is still much to be learned about the nature of this organism, its virulent properties, and its response to host defenses. TB affects the lungs mainly, but can also have other target organs such as brain, spine and the kidneys. When a person with TB infection coughs or sneezes, droplets containing MTB are released into the air and when another person breathes in the infected droplets, they can be infected. However, not everyone infected with TB bacteria becomes sick. There are two TB-related conditions that exist: latent TB (LTB) infection and active TB disease. Those that have latent TB infection do not feel sick or do not present with any symptoms. In LTB cases, even though they are infected with the mycobacterial pathogen, they do not have active TB disease. Overall, it has been shown that about 90% of the people infected with MTB will have LTB infection and 10% will eventually go on to have full-blown active TB at a later stage in their life[5]. Nearly 50% of those who develop TB do so within the first two years of infection. This rate is even higher in immunocompromised individuals, such as those with HIV infection, where the risk of developing TB from LTB activation is significantly higher. Also of particular concern are those infected with drug-resistant TB (XDR TB)[3].
Thus, this ancient human adversary continues to be a challenge in all aspects of medical care, from prevention to diagnosis and therapy.
Currently, there are no tests available to directly detect in vivo the presence of latent MTB in an affected individual and assessment of latent infection involves an imperfect approach of measuring the host immune response to mycobacterial infection[4]. On the contrary, active TB infection is diagnosed by detecting MTB bacteria in clinical samples taken from patients. A positive diagnosis can be made only by culturing MTB from the specimen, even though the results from this may take four to eight weeks for conclusive answers. Other methods for diagnosing TB include chest X-rays, patient sputum smear microscopy, polymerase chain reaction (PCR) testing, immunological memory-based tests including the less specific purified protein derivative (PPD/tuberculin) skin test and more specific Interferon-γ release assays, phage amplification assays, solid and automated liquid cultures, as well as several tests for antibiotic resistance. These tests can only strongly suggest the presence of active TB or LTB as a diagnosis but they cannot confirm the presence of the bacteria in the body. Reliable and rapid diagnosis of latent TB is a major challenge in low socioeconomic areas, and even in parts of developed countries, especially in areas where immunodeficiency diseases like acquired immune deficiency syndrome are more endemic. In many cases, patients have to undergo time-consuming multiple testing before reaching an apparent diagnosis. This testing deficiency can be especially critical when trying to identify high-risk individuals for prophylactic regimen, and also for identifying and managing extrapulmonary TB sites in HIV co-infected patients[6].
Treatment for active TB cases consists of a combination of four first-line antibiotics for a period of two months, followed by two drugs for another four months[7]. First line antibiotics consist of rifampicin, isoniazid, pyrazinamide, and ethambutol. These drugs are effective mainly in actively dividing bacilli and its effectiveness in treating LTB, where the bacilli are dormant, has not yet been proven. The treatment of LTB is usually long term with the intent of sterilizing the non-replicative or slowly replicating bacteria[8]. If the particular mycobacterial strain is resistant to the first line drugs, then treatment is escalated for up to 18 mo with five lines of available drugs. Surgery is also performed as a last resort if treatment fails due to drug resistance. Since the chemotherapy regimen for active and latent TB infections usually spans many months, poor patient compliance rates is a major issue contributing to the emergence of resistant strains[4]. There is a pressing need for rapid and inexpensive tests to confirm latent TB cases in order to manage this global epidemic.
Developing a direct MTB imaging screening tool for the asymptomatic population along with novel treatment strategies is vital to our fight against TB. This is especially true for high-risk categories with LTB such as drug users with unsanitary needles, healthcare workers in high risk and densely populated environments, the medically under-served poor and minority populations, children exposed to high risk adults, immunocompromised patients and patients on immunosuppressant drugs, and health care workers who serve these high risk populations[9]. Developing a direct MTB imaging screening tool with combined therapeutic applications for the asymptomatic population is going to have vitally important and far reaching impact in the fight against TB.
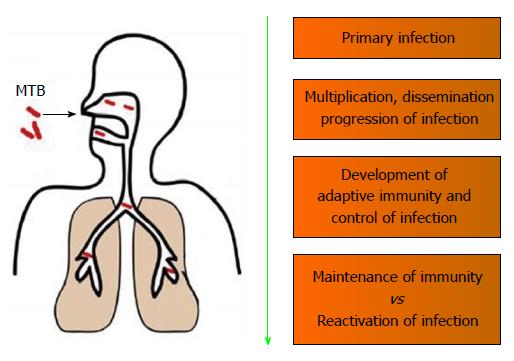
Humans are the only natural host of MTB and are highly susceptible to MTB infections. Even a few (5-10) bacilli is capable of mediating a primary infection[10]. The initial interaction of the MTB with the host involves alveolar macrophages, which is the only known cell type to harbor MTB in vivo[11]. Upon coming in contact with MTB, the interaction of the host immune response with MTB can be divided into 4 general types of events[12]: (1) Primary infection event involving the invading MTB; (2) Events that would promote the dissemination and progressions of the MTB infection; (3) Development of an adaptive immunity that would lead to the containment MTB infection; and (4) Interplay of protective immunity involved in latency vs immunologic compromises leading to reactivation of MTB infection (Figure 1).
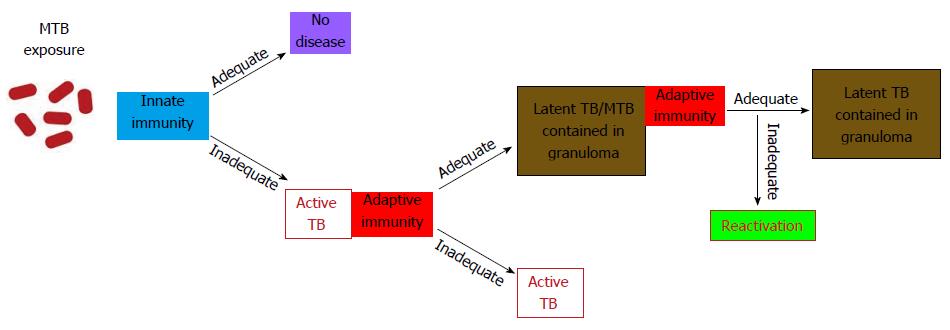
Phagocytosis of the MTB by alveolar macrophages followed by its intracellular growth initiates the cascade of immune events of the primary infection[13]. Activation of the components of the innate immunity, the recruitment of various classes of monocytes and lymphocytes to the site of infection, and the final development of specific immunity allow for the containment of infection (Figure 2).
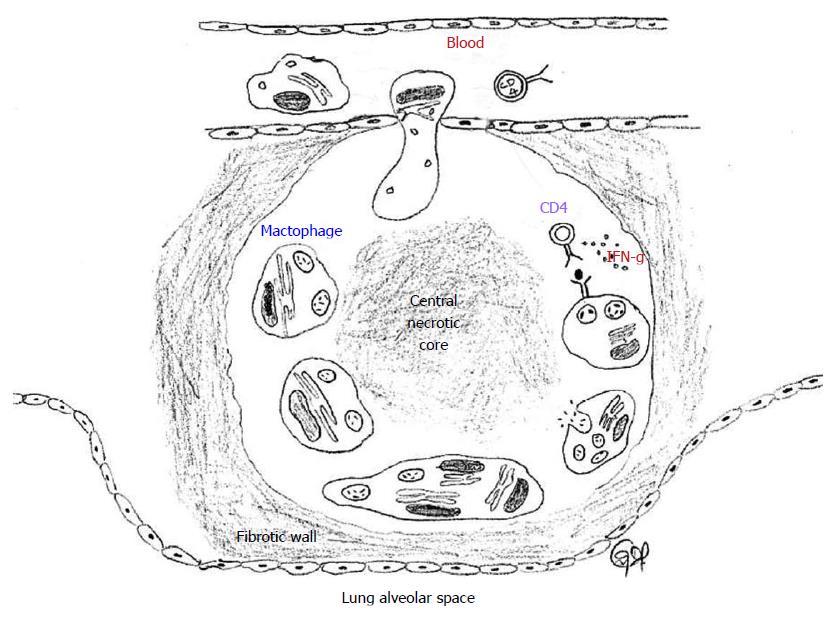
The hallmark of latent TB is the granulomatous lung parenchymal lesions and their draining lymph nodes which is called the “Ghon complex”. The events leading to the formation of granuloma begins when the MTB is inhaled into the lungs and the bacterium is phagocytosed by alveolar macrophages and dendritic cells. The infected cells release proinflammatory cytokines that help recruit more immune cells to the site of infection. The cytokines IL-12 and IL-18 from the infected cells induce natural killer cell activity, which in turn produce IFN-γ that help active macrophages to produce tumor necrosis factor-α and other micobicidal substances. Through the actions of these cytokines and chemokines, other immune cells are recruited leading to the formation of the granuloma[14,15]. In the granuloma, the macrophages further differentiate into epitheloid cells and foamy macrophages and are surrounded by lymphocytes and an outer layer of fibroblasts and matrix proteins. The morphology of the lung granuloma is characterized by a central necrotic core surrounded by concentric layers of macrophages, epitheloid cells, multinucleated Langhans giant cells, and lymphocytes[16,17]. Containment of MTB at the site of primary infection by a cellular wall and a fibrotic outer layer prevents the pathogen from dissemination throughout the host and focuses the immune response to the site of mycobacterial persistence (Figure 3). Successful containment of the pathogen to the site of the primary lesion results in latent infection, which appears in chest X-rays as calcified granulomatous lesions[18].
The exact location of dormant MTB organisms in latent TB has not been elucidated[18]. Studies have shown evidence of the presence of the pathogen in the normal tissue surrounding the granuloma necrotic centers[19] which appears to be the preferred location of the pathogen during latency[20].
A dynamic balance between the host immune response and the MTB pathogen is maintained during latency. Direct cross-talk between MTB and the host immune response occurs in a dense region surrounding the granuloma which is derived from lymphocyte infiltration[21,22]. The granuloma has a central necrotic core which serves as nutritional source for persisting mycobacteria, surrounded by the thick leukocyte wall which prevents the spread of the mycobacteria. The leukocyte derived wall surrounding the granuloma is highly vascularized and facilitates the delivery of drugs against latent TB[23].
Once entering the host, the ability of MTB to survive decades within the body of the host by subverting the host immune defenses is of continued intrigue and fascination. The precise mechanism of how the bacteria is able to achieve this long term dormancy leading to LTB is still unknown, however, recent advances in mycobacterial molecular biology have shed some light into these processes.
Macrophages make up the major component of the innate host defense, and they do this by pathogen recognition, ingestion and killing of foreign microbes that enters the body including pathogenic and non-pathogenic mycobacteria. The pathogenic mycobacteria have developed a number of strategies to subvert the host immune defenses and evade the destructive action of the macrophages, eventually surviving within this normally inhospitable cell for long periods of time thus resulting in the disease[24]. The surviving bacteria within the macrophages can be in a latent state with stationary growth or, given the right conditions in an immunocompromised host, can switch to a metabolically active state that facilitates proliferation, dissemination and active disease.
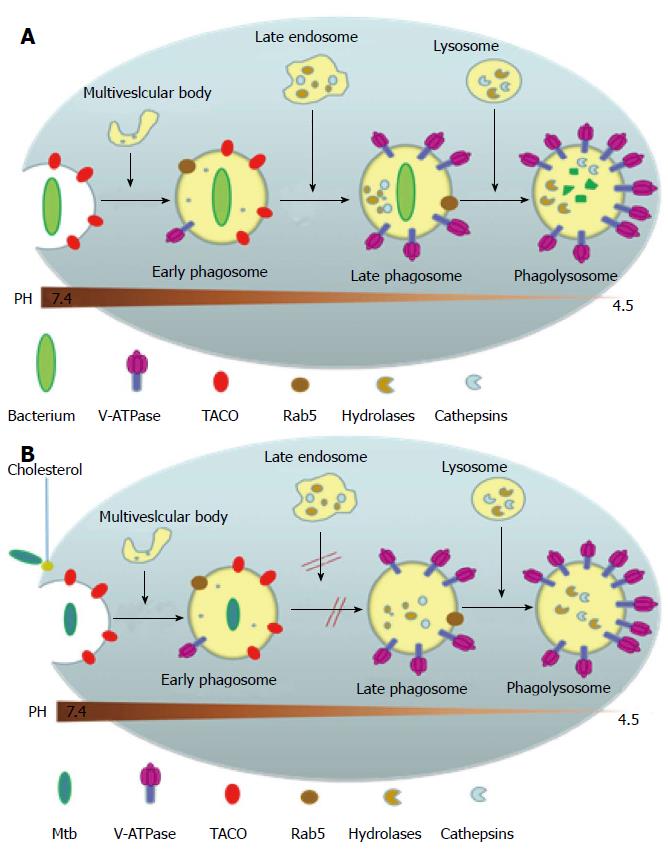
Upon gaining entry into the body, most non-pathogenic microbes get phagocytosed by the macrophage into a phagosome where the invading microbe gets exposed to high levels of reactive oxygen species and reactive nitrogen species. The phagosome then goes on to mature and fuse with the organelles of the endocytic pathway, thereby acquiring surface molecular markers which leads to the acidification of the phagosome to pH 5 as well as gaining hydrolytic enzymes that digest the invading microbe[25,26] (Figure 4A). MTB, however, has developed several ways to evade attack by the macrophage and creates a favorable environment for replication (Figure 4B). This is mainly by inhibiting several aspects of phagosomal maturation, including fusion and fission events along the endocytic pathway and the recruitment of vacuolar H+-ATPases[27,28]. The MTB carrying phagosome retains characteristics of an early phagosome with regard to its pH (about 6-6.5), presence of Rab5 (a Rho-GTPase directing endosomal trafficking and mediating fusion between phagosomes and other organelles), and continued access to other recycling endosomes[29,30], but it lacks mature hydrolases[31] and cathepsins, with interactions between the phagosome and the trans Golgi Network blocked[32]. Several MTB products are believed to be inhibitors of phagosomal maturation, including components of the mycobacterial cell envelope such as lipoarabinomannan, trehalose dimycolate and sulpholipids, phosphatase SapM and kinase PknG[29], and the secreted protein ESAT-6[33]. The exact mechanism behind the inhibition of phagosomal maturation by the mycobacteria is yet to be elucidated.
Studies have shown that cholesterol is a necessary component for the uptake of the MTB into the macrophage via the complement receptors and for the inhibition of phagosomal maturation[34] (Figure 4B). A host protein associated with the cell membrane called tryptophan-aspartate containing coat protein (TACO) is recruited and retained in the phagosomes harboring mycobacteria thereby preventing the bacterial delivery to lysosomes[35]. TACO is an actin-binding protein seen associated with cholesterol rich regions of the host macrophage plasma membrane[34]. The mycobacterium within the phagosome is somehow able to prevent the removal of the TACO coat protein which prevents the fusion of phagososome with lysosome[35]. Moreover, it has been shown that TACO-mediated uptake of mycobacteria depends on cholesterol[34,36,37]. The mycobacterial proteins and genes mediating these adaptive responses inside the macrophage is largely yet to be determined. MTB’ unique ability to utilize cholesterol, a component of cell membranes, also plays a role in its persistence[38]. In the nutrient-deficient intracellular environment, MTB adapts its metabolism by alternating between carbohydrate and fatty acid metabolism[39]. Studies have shown that MTB utilizes cholesterol for its energy needs and for the biosynthesis of virulence-associated lipid phthiocerol dimycocerosate[38]. A number of reports indicate that MTB metabolizes cholesterol during host infections and the metabolic products contribute to the long-term survival of MTB in the host[38,40,41]. Furthermore, because the cholesterol catabolism pathway requires a large number of oxygenases, it should be no surprise that MTB infects the lungs where oxygen concentration is the highest[42].
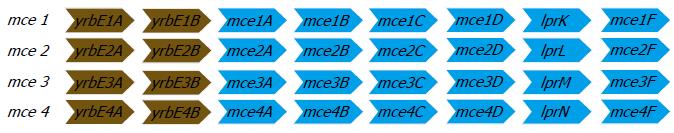
Over the recent years, it was shown that a DNA fragment from MTB cloned into Escherichia coli (E. coli) could mediate the latter’s entry and survival in mammalian cells[43] and was named as the mammalian cell entry (mce) operon. The mce operon genes have been shown to be important in the invasion of the mammalian host cell by the mycobacterium and for establishing a persistent infection both in vitro and in mouse models[44,45]. The analysis of the complete genome sequence of MTB in 1998[46] showed that the mce operon is composed of a group of four homologous mce operons (mce1, mce2, mce3, and mce4). It was found that all the constituent mce genes in the four operons were arranged in an identical manner. Each of the operon contained eight genes, of which two genes preceding the mce genes are named yrbEA and yrbEB which encoded for integral membrane proteins and the six mce genes potentially encoding exported proteins (secreted or surface-exposed) thought to be important for the entry and survival of the pathogen in the mammalian cells[46]. The four mce operons are widely seen throughout the genus Mycobacterium[34] and the general organization of the genes in each of the four operons are shown in Figure 5.
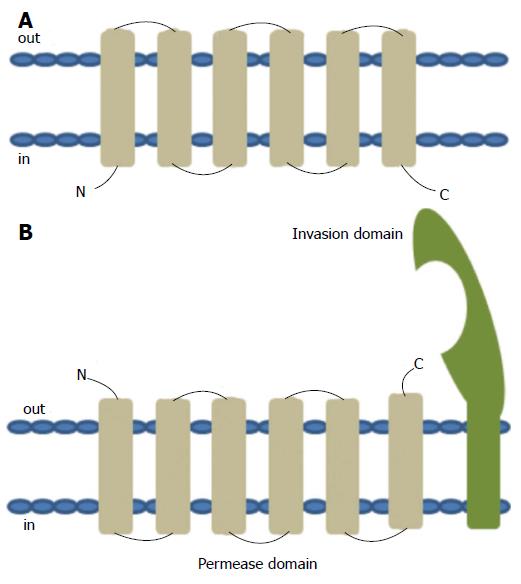
The YRBE and MCE proteins encoded by the MTB mce operons have structural homology to the permeases and the surface binding protein components of the ABC transporters, respectively[47]. The typical ABC permease contains six trans-membrane helices with the C-terminus located on the cytoplasmic side of the membrane (Figure 6A). The YRBE permease contains five or six transmembrane segments outside the C-terminus and the orientation of the N-terminal transmembrane helix may be either cytoplasmic or outside (Figure 6B), suggesting a transmembrane transport role[47,48].
The MCE protein, including MCE1A, 3A and 4A, but not MCE2A, make up a patch of 275-564 amino acid residues, with the hydrophobic stretch at the N-terminal anchored in the membrane, after folding and modification[47], along with a 22 amino acid “invasion domain” near the C-terminal exposed outside the membrane[49,50] (Figure 6B). These characteristics are consistent with their cell surface location and proposed role in cell invasion and immunomodulation[51], however, the mechanism of interaction between YRBE and MCE proteins is not yet clear[52].
Phylogenetic studies have shown that the MCE proteins share between 30%-70% amino acid identity to their inter-operon counterparts but only 16%-26% identity with other MCE proteins encoded by the same operon. For example, there is 61% identity between MCE1A and MCE2A, however, it falls to 25% identity between MCE1B and MCE1C[53,54]. Because of the multiple mce operons in the genome, it is proposed that they may have redundant or time dependent activities. This possibility is supported by the temporal transcriptional expression differences during different stages of in vitro growth between mce1 in comparison to mce3 and mce4[55]. Differences are also seen with in vivo growing bacilli in that even though mce1, mce3, and mce4 transcripts are detectable up to 24 wk post infection in rabbit lung tissue, only mce4 transcript is detectable 16 wk post infection in guinea pig spleen[55]. The expression of mce2 was not detected under any conditions tested[55].
Studies using mce operon deletion or disruption mutants of MTB have demonstrated varying effects with the different mce operons. Some studies have shown than disrupting mce operons lead to attenuation[56,57], while others have shown some degree of hypervirulence for the host MTB following the mutations[58,59]. Deletion of mce operons 3 and 4 attenuated MTB virulence in infected macrophages[60].
The mce4A gene is the first among the six mce genes in the mce4 operon that is studied the most. Studies showed that the MCE4A protein is not only important for host cell invasion but also for survival of the MTB pathogen in human macrophages[61,62]. Individual mce1, mce2, mce3, and mce4 mutants administered intranasally or intravenously in mice have shown to result in lower bacterial burdens and slower mortality of the infected mice, with mce4 operon deletion showing the greatest effects on MTB virulence[56,60]. The route of infection was also shown to be having an effect on the attenuation results in one study[56]. Hypervirulence among mce1 mutants have been demonstrated in two separate studies when administered intranasally, intravenously, or intraperitoneally[58,59]. The deletion mutants in all the above studies, however, were not identical in the nature of their deletions and had variations in their deletion sequences that possibly led to different polar effects on downstream genes, which may explain the discrepancy in the results of some of the deletion mutant studies.
Apart from its possible role in mediating host infection, it is thought that at least some of the MCE4 proteins form an outer membrane channel that mediates cholesterol entry into the cell, thereby enabling uptake by the mycobacterium of host lipids vital for its survival during the prolonged latent infection[63]. Transposon Site Hybridization studies have shown that certain MTB genes involved in the lipid metabolism was genetically linked to the mce4 operon genes[60]. The mce4 has been shown to encode a cholesterol transport system that enables the mycobacterium to derive both carbon and energy from the host membrane lipids and also for possibly generating sterol metabolites mediating its long-term survival within the macrophage[38], with the mce4 expression progressively increasing as the latency phase advances[64].
Hundreds of bacterial encoded short interfering RNAs (siRNA) have been reported over the past decade[65-67]. Majority of these siRNAs act by binding to their target mRNAs to bring about the repression. They fall into two major categories: some are encoded at locations farther away from the target gene (trans-acting) and others are encoded by DNA strand complementary to the target gene (cis-acting). The trans-acting siRNAs generally share only limited complementarity with their target gene and thus is prone to have off target effects. Trans-acting siRNAs are by far the most characterized bacterial siRNAs and have been shown to usually require the chaperon protein Hfq for base pairing[68]. The cis-encoded siRNAs, or anti-sense RNAs, have perfect complementarity with their target gene and thus have more extensive and stronger base pairing. Among the reported bacterial antisense RNAs, some are short (siRNA), with around 100 nucleotides in length and are usually encoded by plasmids or bacteriophages, while some are chromosomally encoded and are longer, in some cases overlapping entire genes or corresponding to the 5’ or 3’ extension of the protein coding region of the mRNA. The 5’ untranslated region of the mogR mRNA in Listeria monocytogenes overlaps 3 genes on the opposite strand involved in the flagellar synthesis and serves as an example of chromosomally encoded long antisense RNAs[69]. The binding region of antisense RNA on the target mRNA can also vary and may be located in the 5’ end, 3’ end, the central region, or the entire coding region.
Antisense RNAs in the bacterial cell have been shown to repress many detriments to the cell such as transposons and toxic proteins. One of the first antisense RNA to be discovered in bacteria was the RNA-OUT of the transposon Tn10, which was shown to inhibit transposition by preventing the translation of transposase mRNA[70]. Antisense RNAs are also seen encoded opposite transposase genes in Salmonella enterica[71,72], Caulobacter crescentus[73], and Listeria monocytogenes[69]. Thus a critical role of bacterial antisense RNA, as in eukaryotes[74], appears to be the inhibition of transposition. There is increasing evidence that antisense RNAs downregulate the expression of toxic proteins[75,76]. It has been found that most of these repressed proteins are hydrophobic, small with less than 50 amino acids, and toxic at higher levels. An example of this tight repression of one such toxic protein is seen in E. coli, where the low levels of SymE protein is maintained by the LexA repressor of the save our souls response, the SymR antisense RNA and the Lon protease[77]. Some of the antisense RNAs to transposases and toxic genes, such as SymR, are expressed constitutively in the cell[77].
Studies have shown that antisense RNAs can positively and negatively regulate the expression of various transcriptional regulators and other metabolic and virulence proteins in bacterial systems. For example, The 109 nucleotide GadY antisense RNA of E. coli overlaps the intergenic region of the dicistronic gadXW mRNA which encodes two transcription regulators of the acid stress response genes and enhanced transcription of the GadY gene which leads to cleavage of gadXW mRNA into gadX and gadW transcripts, leading to positive regulation (increased expression) of those genes[78,79]. And on the other hand, in the nitrogen-fixing cyanobacterium Anabaena sp. PCC 7120, the approximately 2200 nucleotide alr1690-α-furA antisense RNA spans the entire alr1690 coding region and extends through the gene encoding the ferric uptake transcriptional regulator, FurA, into its promoter and regulator regions and it helps decrease furA expression and translation, thereby acting as a negative regulator of iron absorption and nitrogen metabolism[80]. Similar regulatory RNAs controlling metabolic responses to environmental effects have been reported in many other bacterial systems[81-87].
Another recently discovered phenomenon is the antisense-mediated gene regulatory switch in the bacteria called the “excludon”. This comprises a gene locus encoding an unusually long antisense RNA that spans divergent genes or operons with related or opposing functions. In such a regulatory system, the antisense RNA can inhibit the expression of one operon while functioning as an mRNA for the adjacent operon, there by acting as fine-tuning regulatory switches in bacteria[88].
Antisense RNA also regulates the expression of various structural and virulence factors in different bacteria. For instance, the 1200 nucleotide AmgR RNA which is encoded opposite the Salmonella enterica mgtCBR operon is responsible for the bacteria’s virulence and survival in macrophages[89]. A number of other antisense RNAs modulating virulence and regulating host-pathogen interactions have been discovered in a variety of bacterial species over the years[90-97]. Antisense RNAs have also been found to impact other benign structural components including flagellar synthesis in Rhizobium[98], H. pylori[99], L. monocytogenes[69] and S. enterica[100].
The first complete experimental confirmation of short antisense RNAs in mycobacteria was published in 2009, which revealed 5 trans-acting and 4 cis-acting siRNAs in MTB H37Rv in the context of pH and oxidative stress[101]. By the end of 2013, a total of more than 200 endogenous antisense RNAs were experimentally identified in various mycobacteria, including 70 in MTB[102-110], 90 in M. Bovis[103,104,111], 9 in M. avium[112], and 44 in M. smegmatis[102-104,113]. From these recent studies, a stronger connection between mycobacterial pathogenesis and the levels of expression of the antisense RNAs have emerged but many new questions about their potential pathogenic vs housekeeping functions remain to be answered. The lack of identification of an Hfq homolog in mycobacteria prevents the current approach of coimmunoprecipitation, making the study of the role of antisense RNAs all the more difficult in this genus[102]. Pandey et al[114] have proposed an alternative protein, Rv2367, as a potential RNA chaperon in place of Hfq[114], however, studies are ongoing in this direction to find a functionally-equivalent chaperon or to get around this issue[113]. Also, the role of mycobacterial antisense RNAs in regulating transposition is not as clear as in other bacterial systems like E. coli.
Antisense RNAs can repress or modulate expression of target genes by a variety of mechanisms, including transcription interference, transcription attenuation, degradation by endo- or exonucleases, or by blocking ribosome binding. When inducing transcription interference, the transcription of antisense RNA from one promoter hinders the RNA polymerase from either binding or extending the target gene transcript from the opposite strand[115,116]. This type of interference occurs only in Cis and does not involve base pairing. In transcription attenuation, the binding of the antisense RNA to the target RNA causes a conformational change creating a terminator structure in the mRNA leading to its premature termination of transcription[117]. Antisense RNAs can affect target mRNA stability by stimulating or inhibiting its degradation. When employing endo- or exonucleases for gene regulation, the antisense RNA, upon binding to its target mRNA, induces or blocks a ribonuclease target site within the mRNA or can indirectly block the binding of the ribonuclease at a distant site. In many bacterial systems, two major endoribonucleases have been identified, RNase III which cleaves double stranded RNA into two with different stabilities than the original transcript[118-121], and RNase E, a component of the multi-protein degradasome complex which cleaves single stranded RNA and interacts with Hfq and globally affect mRNA stability[119,122]. It is not precisely clear how the antisense RNA modulates RNase E activity, but the proposed mechanisms include the donation of its 5’ monophosphate to stimulate RNase E activity or physically block the RNase E recognition site by basepairing to downregulate activity[123]. Other ribonucleases in bacteria have also been identified with more specialized functions, including RNase G (a non-essential paralog of RNase E)[124,125], RNase P[126,127], RNase LS[128], RNase Z[129,130], RNase H[131], RNase J1/J2[132], and the recently characterized RNase Y[133]. Many of these ribonucleases have already been characterized in various Mycobacteria, including MTB and M. smegmatis[134-142]. Apart from these mechanisms, it has also been found that some antisense RNAs can physically block mRNA expression by binding to the Shine Dalgarno sequences of their target mRNA and prevent ribosome binding[66,77,80], or they may indirectly modulate expression by altering the target mRNA conformation[67]. Finally, antisense RNAs can exhibit dual functions by acting as mRNAs and antisense RNAs or cis- and trans- acting RNAs[78].
Two general mechanisms have been proposed for base-pairing in antisense RNAs. The first type is a single-step mechanism in which the antisense RNA makes initial contact with the target mRNA to form a duplex[143]. The second type is a multi-step system in which the initial duplex formed is stabilized by a protein, followed by the formation of the more stable complete duplex[144,145]. In many cases of base pairing for the antisense RNA, a stem-loop structure is found to be important, along with a “pyrimidine-uracil-any nucleotide-purine” U-turn motif[146].
Synthetic antisense RNAs are generally delivered either by expressing the antisense transcript from a gene introduced into the cell or by direct delivery of antisense oligonucleotides. Degradation of the antisense transcripts can be a problem for both these delivery approaches, however, this issue is mitigated by using sequences that form more stable hair-pin structures with paired ends[147]. In order to increase the stability and uptake, antisense RNAs have been modified in many ways, including the addition of peptide nucleic acid (PNA) or alternating 2’O-methyl to their backbones, switching ribose rings to morpholine rings (PMO), switching internucleoside bonds with phosphorothioates (PS-ODNs), or by conjugating cationic peptides to PNAs and PMOs[148-150].
Antisense technology using synthetic siRNAs have been used as a powerful tool in knocking down genes in prokaryotes (and also in eukaryotes), including hepatitis G virus[151], influenza virus[152], picornavirus[153], and Trypanosoma brucei[154]. When targeted to essential genes, siRNAs inhibit growth of E. coli[148,149,155,156], S. enterica[157], Staphylococcus aureus[158], M. smegmatis[159] and MTB[61,160]. Antisense RNAs have been successfully used for the study of bacterial growth and metabolism since this approach allows conditional knock down of target genes[161-165]. It has also been used for the study of various putative virulence factors in bacteria[166,167]. Antisense technology has helped in identifying new antibiotics[168,169], antibiotic targets[170,171], sensitizing bacteria to antibiotics[171-173], and to elucidate the mechanism of action of potential new drugs[171].
Molecular beacons are a newer class of antisense RNA tagged with a fluorophore/quencher pair and their use for in vivo detection and knockdown of mRNA is gaining popularity. Molecular beacon based short interfering RNA (MB siRNA) has recently been proven to be a powerful tool for therapeutic gene silencing because of its specificity, broad applicability, and high efficiency[174-176]. The on/off signals produced by the fluorophore/quencher pair depends on the conformational state of the MB (Figure 7). In the absence of the target mRNA, the stem brings the quencher in close proximity with the fluorophore and turns the fluorescence off with high quenching efficiency via fluorescence resonance energy transfer (FRET). In FRET, the energy from the donor chromophore is transferred to acceptor quencher near-by thus resulting in the absence of fluorescence. If the quencher and the fluorophore are far apart (following hybridization of the beacon to its target) then the quencher molecule will not be able to absorb the energy from the donor fluorophore. This would result in an increase in fluorescence. This technology has been used to detect mRNA expression in cells as well as for the detection and knockdown of telomerase expression in human breast cancer cells[177], BMP4 mRNA in hedgehog signaling[178], aromatase mRNA in breast cancer cells[179], and for the detection and attenuation of mce4a mediated M. smegmatis infection in macrophages as discussed in section below[155,180].
The mce operons are widely seen throughout the genus Mycobacterium and a homolog of mce4 has been confirmed in the mycobacterial species M. smegmatis[64,181]. Even though M. smegmatis is non-pathogenic, previous studies have shown that it can survive and multiply within macrophages in a pathogen-like manner by manipulating the host cell during initial stages by delaying phagosomal acidification and recruitment of V-ATPase[28,182], thus making it a suitable model to study mce4 operon mediated invasion and intracellular mycobacterial survival.
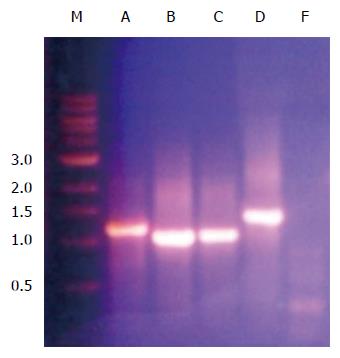
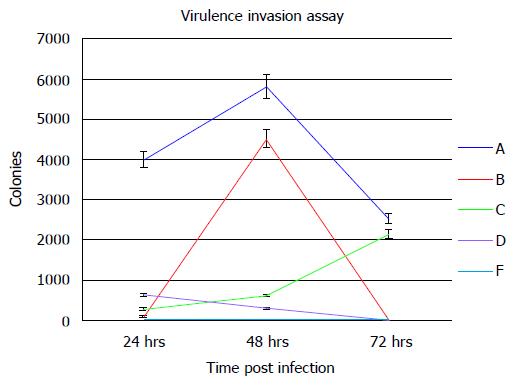
In order to determine the ability of a siRNA molecular beacon to detect and attenuate mycobacterial infection in macrophages, we first conducted experiments towards determining the most infective gene in the mce4 operon. Because of the slow growth rate of MTB and also due to the high degree of homology between mce4 operons of mycobacteria[181], the mce4 operon of the rapid growing M. smegmatis was selected for our studies. Using gene specific primers with the reverse primer for each set excluding the termination codon, each of the mce4 genes, mce4A, mce4B, mce4C, mce4D, and mce4F were PCR amplified, cloned into the prokaryotic expression vector pTrcHis2-TOPO and stably expressed in E. coli. Western blot analyses with monoclonoal antibodies against c-myc and 6xHis showed that the MCE4 proteins were expressed in host E.coli. Next we conducted invasion assays in MCF7 breast cancer cells using E. coli clones expressing the M. smegmatis genes and the results showed that mce4A-F conferred virulence to its host E.coli. However, mce4A appeared to confer the earliest virulence to its host E. coli and the virulence was found to be sustained during the entire invasion period (72hr)[180]. We later, repeated the cloning experiments using the mce4 operon genes of MTB, by PCR amplifying each of the mce4A-F (Figure 8), cloning into TOP10 E.coli using the prokaryotic expression vector pTrcHis2-TOPO, and performing invasion assay using MCF7 breast cancer cells. Our results showed that, as with M. smegmatis, the mce4A gene conferred the greatest degree of virulence to its host E. coli (Figure 9). Therefore, mce4A was selected as the target gene for designing a molecular beacon antisense RNA.
The mce4A antisense molecular beacon RNA was designed to have a stem-loop structure, with the nucleotides in the stem complementary to each other to form a 5-base pair double stranded stem and the loop consisting of 20 nucleotides that are complementary to a region of the target mce4A mRNA in M. smegmatis. Also, conjugated to the 5’ and 3’ ends of this molecular beacon are the fluorophore 5’ TYE 665 and quencher Iowa Black RQ-SP (fluorophore quencher for 500-700 nm spectrum), respectively. This molecular beacon design combined both detection and therapeutic capabilities[177-179]. The rationale is that in the absence of the target mce4A mRNA, the molecular beacon remains in its hairpin form while in the presence of its target mRNA the 20 nucleotide loop will compete with the 5 nucleotide stem for hybridization to their target mce4A mRNA and the stem to its complementary pairs on the opposite ends of the target sequence. The hybridization potential of the loop to its target, based on the number of nucleotides within it (20 vs 5), will be greater than that of the strands for the stem. Hybridization of the loop to the mce4A mRNA will separate the fluorophore from the quencher thus inducing fluorescence (detection) and degradation (therapeutic) of the mRNA. Since the mycobacterium utilizes the product of mce4A for entry in to macrophages and for its survival using host cholesterol for carbon and energy source transported through the MCE4 transporters[41,45,62,183], the degradation of the mce4A mRNA will lead to its reduced survival. Our studies first tested the ability of the mce4A siRNA to detect its target mce4A mRNA in M. smegmatis and in macrophages infected with M. smegmatis and the results show that the molecular beacon siRNA detected its target in M. smegmatis and in macrophages infected with M. smegmatis. Thus, we were able to show that a molecular beacon can be designed against one of the mce4 operon genes in M. smegmatis that facilitates the detection of mycobacterial infection in macrophages.
Tests were carried out to test the ability of this siRNA molecular beacon to not only detect but also attenuate mycobacterial infection in macrophages. Towards this end, we used a green fluorescent protein (GFP) expressing lentiviral vector, piLenti-siRNA-GFP, to successfully transduce and stably express the mce4A siRNA molecular beacon construct in macrophages infected with either E. coli expressing mce4A gene (E. coli-4A) or M. smegmatis. Using confocal imaging and Western blot analyses with anti-GFP antibodies, we were able to demonstrate stable expression of siRNA up to 48 h post transduction and infection using the GFP reporter.
After confirming the expression of the GFP protein by fluorescence imaging and Western blot analyses, invasion assay was carried out to assess the effect of mce4A siRNA on mycobacterial infection in macrophages. For this, differentiated U937 macrophages were transduced with piLenti-siRNA-GFP phage for 24hrs followed by infection with E.coli-4A or M. smegmatis for 3 h, and incubation for 0, 3, 6, 24, and 48 h, respectively. The cells were extensively washed and lysed in 0.1% Triton-X 100 lysis buffer and the lysates were plated on either LB agar containing 100 μg/mL ampicillin for E. coli-4A or 7H11 media for M. smegmatis. The degree of attenuation of E. coli-4A infection was compared between 3, 6, 24, and 48 h against that at 0 h baseline and was found to be 0%, 77%, 59.6%, and 99.7%, respectively. The degree of attenuation of M. smegmatis infection was compared between 3, 6, 24, and 48 h against that at 0 h baseline and was found to be 94.8%, 70.3%, 98.9%, and 93.4%, respectively. Thus, our results showed that the mce4A siRNA was able to significantly attenuate both E. coli-4A and M. smegmatis infection in macrophages[159].
Separate set of experiments were conducted to further test the hypothesis that the mce4A siRNA molecular beacon can attenuate the mce4A mRNA levels in E. coli expressing mce4A gene within infected macrophages. For this, reverse transcription polymerase chain reaction analysis was performed on lysates from differentiated U937 cells which were transduced with the piLenti-siRNA-GFP phage for 24 h, followed by infection with E. coli-4A for 3 h and incubation for 0, 3, 6, and 24 h. The cells were washed and lysed and the intracellular bacteria were isolated and washed at each time point of incubation. The bacterial sample from each of the time points were lysed and the mRNA was isolated and purified using DNAse 1 enzyme treatment. Reverse transcripts were generated using RTPCR and the cDNAs were amplified using gene specific primers for M. smegmatis mce4A and E. coli 16S rRNA gene as internal control. The degree of attenuation of mce4A mRNA levels was compared between 3, 6, and 24 h against that at 0hr and the results were found to be 0%, 81%, 40%, and 36%, respectively using densitometry gel analysis. Our results thus showed that mce4A siRNA was able to attenuate mce4A levels within infected macrophages as opposed to E. coli 16S rRNA internal positive control and the degree of attenuation of mce4A mRNA levels in E. coli-4A was found to be significant.
Thus, we have successfully demonstrated that a molecular beacon can be designed against one of the mce4 operon genes in M. smegmatis which can be used to both detect and attenuate mycobacterial infection in macrophages.
Antisense oligonucleotides, considered the pharmacology of the future[184], interact with their mRNA targets with greater specificity and binding affinity than traditional drugs to their protein targets. Recent advances have enhanced their hybridization to target mRNA, reduced their overall toxicity with decreased susceptibility to cellular nucleases. The lung provides an excellent target for direct antisense oligonucleotide delivery by inhalation, thereby achieving a bolus dose directly to the target site. Cationic lipids in the lung surfactants enhance oligonucleotides entry into cells[185]. Penetration of the inhaled oligonucleotides into deeper tissues of the lung has been established by autoradiogram, surgical dissection and receptor quantification studies[186]. Further studies to test the hypothesis that mce4 siRNA respirable molecular beacons can localize and attenuate mycobacterial infection in pulmonary granulomas in animal models will take the fight against TB a long way in eradicating this versatile human pathogen.
The association of the mce operons, especially that of mce4, with mycobacterial invasion and latency is no longer considered casual and with strong evidences emerging over the recent years it can now be considered as a potent mediator of MTB infection and survival in its only human host. The mce invasion domain is equipped to mediate the entry and localization of the bacteria in the host macrophages at cholesterol rich regions creating cholesterol-associated protein coated phagosomes, thereby creating an ingenious mechanism for subverting the immune defenses. Another paradigm to the mycobacterial saga was added by the discovery that the MCE associated protein, YRBE4 transporters, in conjunction with the MCE4 domains, transport cholesterol into the cell for its energy and carbon needs, which then possibly generates metabolites that can further mediate its latency in the host. Strategies like identifying the level of infectivity of individual mce operon genes and designing efficacious drugs like molecular beacon siRNAs against mce targets can aid in the simultaneous detection and eradication of this elusive human pathogen.
| 1. | WHO. Tuberculosis global facts 2010/2011. Cent Eur J Public Health. 2010;18:197. [PubMed] |
| 2. | Eurosurveillance editorial team. WHO publishes Global tuberculosis report 2013. Euro Surveill. 2013;18. [PubMed] |
| 3. | Centers for Disease, C, Prevention. Trends in tuberculosis--United States, 2012. MMWR Morb Mortal Wkly Rep. 2013;62:201-205. [PubMed] |
| 4. | Horsburgh CR, Rubin EJ. Clinical practice. Latent tuberculosis infection in the United States. N Engl J Med. 2011;364:1441-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 205] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 5. | Kumar V, Robbins SL. Robbins basic pathology (8th ed.). Philadelphia, PA: Saunders/Elsevier 2007; . |
| 6. | Chakraborty MS, Chakraborty A. Tuberculosis and HIV illness. J Indian Med Assoc. 2000;98:103-106, 109. [PubMed] |
| 7. | Caminero JA, Sotgiu G, Zumla A, Migliori GB. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis. 2010;10:621-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 370] [Cited by in RCA: 381] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 8. | Ehlers S. Lazy, dynamic or minimally recrudescent? On the elusive nature and location of the mycobacterium responsible for latent tuberculosis. Infection. 2009;37:87-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Griffith DE, Kerr CM. Tuberculosis: disease of the past, disease of the present. J Perianesth Nurs. 1996;11:240-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Smith DW, Wiegeshaus E, Navalkar R, Grover AA. Host-parasite relationships in experimental airborne tuberculosis. I. Preliminary studies in BCG-vaccinated and nonvaccinated animals. J Bacteriol. 1966;91:718-724. [PubMed] |
| 11. | Filley EA, Rook GA. Effect of mycobacteria on sensitivity to the cytotoxic effects of tumor necrosis factor. Infect Immun. 1991;59:2567-2572. [PubMed] |
| 12. | Schlesinger LS, DesJardin LE. Tuberculosis: the microbe host interface. Wymondham, UK: Horizon Bioscience 2004; . |
| 13. | Hossain MM, Norazmi MN. Pattern recognition receptors and cytokines in Mycobacterium tuberculosis infection--the double-edged sword? Biomed Res Int. 2013;2013:179174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 14. | Korbel DS, Schneider BE, Schaible UE. Innate immunity in tuberculosis: myths and truth. Microbes Infect. 2008;10:995-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 179] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 15. | North RJ, Jung YJ. Immunity to tuberculosis. Annu Rev Immunol. 2004;22:599-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 524] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 16. | Boros DL. Granulomatous inflammations. Prog Allergy. 1978;24:183-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Mariano M. The experimental granuloma. A hypothesis to explain the persistence of the lesion. Rev Inst Med Trop Sao Paulo. 1995;37:161-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Ulrichs T, Kaufmann SH. New insights into the function of granulomas in human tuberculosis. J Pathol. 2006;208:261-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 292] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 19. | Feldman WH, Baggenstoss AH. The occurrence of virulent tubercle bacilli in presumably non-tuberculous lung tissue. Am J Pathol. 1939;15:501-515. [PubMed] |
| 20. | Ulrichs T, Lefmann M, Reich M, Morawietz L, Roth A, Brinkmann V, Kosmiadi GA, Seiler P, Aichele P, Hahn H. Modified immunohistological staining allows detection of Ziehl-Neelsen-negative Mycobacterium tuberculosis organisms and their precise localization in human tissue. J Pathol. 2005;205:633-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 21. | Ulrichs T, Kosmiadi GA, Trusov V, Jörg S, Pradl L, Titukhina M, Mishenko V, Gushina N, Kaufmann SH. Human tuberculous granulomas induce peripheral lymphoid follicle-like structures to orchestrate local host defence in the lung. J Pathol. 2004;204:217-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 250] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 22. | Fenhalls G, Wong A, Bezuidenhout J, van Helden P, Bardin P, Lukey PT. In situ production of gamma interferon, interleukin-4, and tumor necrosis factor alpha mRNA in human lung tuberculous granulomas. Infect Immun. 2000;68:2827-2836. [PubMed] |
| 23. | Ulrichs T, Kosmiadi GA, Jörg S, Pradl L, Titukhina M, Mishenko V, Gushina N, Kaufmann SH. Differential organization of the local immune response in patients with active cavitary tuberculosis or with nonprogressive tuberculoma. J Infect Dis. 2005;192:89-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 103] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Koul A, Herget T, Klebl B, Ullrich A. Interplay between mycobacteria and host signalling pathways. Nat Rev Microbiol. 2004;2:189-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 286] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 25. | Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111-1122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 767] [Cited by in RCA: 791] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 26. | Denis M. Interferon-gamma-treated murine macrophages inhibit growth of tubercle bacilli via the generation of reactive nitrogen intermediates. Cell Immunol. 1991;132:150-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 259] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 27. | Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1025] [Cited by in RCA: 968] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 28. | Kuehnel MP, Goethe R, Habermann A, Mueller E, Rohde M, Griffiths G, Valentin-Weigand P. Characterization of the intracellular survival of Mycobacterium avium ssp. paratuberculosis: phagosomal pH and fusogenicity in J774 macrophages compared with other mycobacteria. Cell Microbiol. 2001;3:551-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 127] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 29. | Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219:37-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 256] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 30. | Via LE, Deretic D, Ulmer RJ, Hibler NS, Huber LA, Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J Biol Chem. 1997;272:13326-13331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 460] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 31. | Vergne I, Chua J, Singh SB, Deretic V. Cell biology of mycobacterium tuberculosis phagosome. Annu Rev Cell Dev Biol. 2004;20:367-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 339] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 32. | Vergne I, Chua J, Deretic V. Mycobacterium tuberculosis phagosome maturation arrest: selective targeting of PI3P-dependent membrane trafficking. Traffic. 2003;4:600-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 88] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 33. | Tan T, Lee WL, Alexander DC, Grinstein S, Liu J. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol. 2006;8:1417-1429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 129] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Gatfield J, Pieters J. Essential role for cholesterol in entry of mycobacteria into macrophages. Science. 2000;288:1647-1650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 449] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 35. | Ferrari G, Langen H, Naito M, Pieters J. A coat protein on phagosomes involved in the intracellular survival of mycobacteria. Cell. 1999;97:435-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 426] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 36. | Anand PK, Kaul D. Vitamin D3-dependent pathway regulates TACO gene transcription. Biochem Biophys Res Commun. 2003;310:876-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 37. | Kaul D, Anand PK, Verma I. Cholesterol-sensor initiates M. tuberculosis entry into human macrophages. Mol Cell Biochem. 2004;258:219-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 38. | Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA. 2008;105:4376-4380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 722] [Cited by in RCA: 838] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 39. | Muñoz-Elías EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 598] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 40. | Forrellad MA, Klepp LI, Gioffré A, Sabio y García J, Morbidoni HR, de la Paz Santangelo M, Cataldi AA, Bigi F. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4:3-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 354] [Cited by in RCA: 436] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 41. | Miner MD, Chang JC, Pandey AK, Sassetti CM, Sherman DR. Role of cholesterol in Mycobacterium tuberculosis infection. Indian J Exp Biol. 2009;47:407-411. [PubMed] |
| 42. | Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci USA. 2007;104:1947-1952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 419] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 43. | Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261:1454-1457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 287] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 44. | Flesselles B, Anand NN, Remani J, Loosmore SM, Klein MH. Disruption of the mycobacterial cell entry gene of Mycobacterium bovis BCG results in a mutant that exhibits a reduced invasiveness for epithelial cells. FEMS Microbiol Lett. 1999;177:237-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 45. | Senaratne RH, Sidders B, Sequeira P, Saunders G, Dunphy K, Marjanovic O, Reader JR, Lima P, Chan S, Kendall S. Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J Med Microbiol. 2008;57:164-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 83] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 46. | Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5846] [Cited by in RCA: 5881] [Article Influence: 210.0] [Reference Citation Analysis (0)] |
| 47. | Casali N, Riley LW. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics. 2007;8:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 179] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 48. | Liu PQ, Liu CE, Ames GF. Modulation of ATPase activity by physical disengagement of the ATP-binding domains of an ABC transporter, the histidine permease. J Biol Chem. 1999;274:18310-18318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 49. | Das AK, Mitra D, Harboe M, Nandi B, Harkness RE, Das D, Wiker HG. Predicted molecular structure of the mammalian cell entry protein Mce1A of Mycobacterium tuberculosis. Biochem Biophys Res Commun. 2003;302:442-447. [PubMed] |
| 50. | Mitra D, Saha B, Das D, Wiker HG, Das AK. Correlating sequential homology of Mce1A, Mce2A, Mce3A and Mce4A with their possible functions in mammalian cell entry of Mycobacterium tuberculosis performing homology modeling. Tuberculosis (Edinb). 2005;85:337-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 51. | Harboe M, Christensen A, Ahmad S, Ulvund G, Harkness RE, Mustafa AS, Wiker HG. Cross-reaction between mammalian cell entry (Mce) proteins of Mycobacterium tuberculosis. Scand J Immunol. 2002;56:580-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 52. | Zhang F, Xie JP. Mammalian cell entry gene family of Mycobacterium tuberculosis. Mol Cell Biochem. 2011;352:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 53. | Tekaia F, Gordon SV, Garnier T, Brosch R, Barrell BG, Cole ST. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung Dis. 1999;79:329-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 227] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 54. | Ahmad S, Akbar PK, Wiker HG, Harboe M, Mustafa AS. Cloning, expression and immunological reactivity of two mammalian cell entry proteins encoded by the mce1 operon of Mycobacterium tuberculosis. Scand J Immunol. 1999;50:510-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 55. | Kumar A, Bose M, Brahmachari V. Analysis of expression profile of mammalian cell entry (mce) operons of Mycobacterium tuberculosis. Infect Immun. 2003;71:6083-6087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 56. | Gioffré A, Infante E, Aguilar D, Santangelo MP, Klepp L, Amadio A, Meikle V, Etchechoury I, Romano MI, Cataldi A. Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 2005;7:325-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 125] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 57. | Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989-12994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1059] [Cited by in RCA: 1034] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 58. | Shimono N, Morici L, Casali N, Cantrell S, Sidders B, Ehrt S, Riley LW. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc Natl Acad Sci USA. 2003;100:15918-15923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 173] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 59. | Lima P, Sidders B, Morici L, Reader R, Senaratne R, Casali N, Riley LW. Enhanced mortality despite control of lung infection in mice aerogenically infected with a Mycobacterium tuberculosis mce1 operon mutant. Microbes Infect. 2007;9:1285-1290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 60. | Joshi SM, Pandey AK, Capite N, Fortune SM, Rubin EJ, Sassetti CM. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc Natl Acad Sci USA. 2006;103:11760-11765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 144] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 61. | Chandolia A, Rathor N, Sharma M, Saini NK, Sinha R, Malhotra P, Brahmachari V, Bose M. Functional analysis of mce4A gene of Mycobacterium tuberculosis H37Rv using antisense approach. Microbiol Res. 2014;169:780-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 62. | Saini NK, Sharma M, Chandolia A, Pasricha R, Brahmachari V, Bose M. Characterization of Mce4A protein of Mycobacterium tuberculosis: role in invasion and survival. BMC Microbiol. 2008;8:200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 63. | Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H. Mycobacterial outer membranes: in search of proteins. Trends Microbiol. 2010;18:109-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 170] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 64. | Rathor N, Chandolia A, Saini NK, Sinha R, Pathak R, Garima K, Singh S, Varma-Basil M, Bose M. An insight into the regulation of mce4 operon of Mycobacterium tuberculosis. Tuberculosis (Edinb). 2013;93:389-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 65. | McClure R, Tjaden B, Genco C. Identification of sRNAs expressed by the human pathogen Neisseria gonorrhoeae under disparate growth conditions. Front Microbiol. 2014;5:456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 66. | Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol. 2009;160:278-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 111] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 67. | Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615-628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1315] [Cited by in RCA: 1191] [Article Influence: 70.1] [Reference Citation Analysis (0)] |
| 68. | Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 300] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 69. | Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 757] [Cited by in RCA: 705] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 70. | Simons RW, Kleckner N. Translational control of IS10 transposition. Cell. 1983;34:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 251] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 71. | Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913-1927. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 180] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 72. | Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. [PubMed] |
| 73. | Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L. Small non-coding RNAs in Caulobacter crescentus. Mol Microbiol. 2008;68:600-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 79] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 74. | Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656-668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 715] [Cited by in RCA: 635] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 75. | Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743-3759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 215] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 76. | Gerdes K, Wagner EG. RNA antitoxins. Curr Opin Microbiol. 2007;10:117-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 190] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 77. | Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol Microbiol. 2007;64:738-754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 199] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 78. | Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698-6705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 259] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 79. | Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol. 2008;70:965-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 80. | Hernández JA, Muro-Pastor AM, Flores E, Bes MT, Peleato ML, Fillat MF. Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J Mol Biol. 2006;355:325-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 81. | Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci USA. 2006;103:7054-7058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 186] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 82. | Jackson LA, Pan JC, Day MW, Dyer DW. Control of RNA stability by NrrF, an iron-regulated small RNA in Neisseria gonorrhoeae. J Bacteriol. 2013;195:5166-5173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 83. | Massé E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 184] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 84. | Patenge N, Billion A, Raasch P, Normann J, Wisniewska-Kucper A, Retey J, Boisguérin V, Hartsch T, Hain T, Kreikemeyer B. Identification of novel growth phase- and media-dependent small non-coding RNAs in Streptococcus pyogenes M49 using intergenic tiling arrays. BMC Genomics. 2012;13:550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 85. | Recalcati S, Minotti G, Cairo G. Iron regulatory proteins: from molecular mechanisms to drug development. Antioxid Redox Signal. 2010;13:1593-1616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 86. | Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004;32:3340-3353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 139] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 87. | Shioya K, Michaux C, Kuenne C, Hain T, Verneuil N, Budin-Verneuil A, Hartsch T, Hartke A, Giard JC. Genome-wide identification of small RNAs in the opportunistic pathogen Enterococcus faecalis V583. PLoS One. 2011;6:e23948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 88. | Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P. The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol. 2013;11:75-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 130] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 89. | Lee EJ, Groisman EA. Tandem attenuators control expression of the Salmonella mgtCBR virulence operon. Mol Microbiol. 2012;86:212-224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 90. | Shepherd DP, Li N, Micheva-Viteva SN, Munsky B, Hong-Geller E, Werner JH. Counting small RNA in pathogenic bacteria. Anal Chem. 2013;85:4938-4943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 91. | Koo JT, Lathem WW. Global discovery of small noncoding RNAs in pathogenic Yersinia species. Adv Exp Med Biol. 2012;954:305-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 92. | Wadler CS, Vanderpool CK. Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res. 2009;37:5477-5485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 93. | Heroven AK, Böhme K, Rohde M, Dersch P. A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol Microbiol. 2008;68:1179-1195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 99] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 94. | Postic G, Dubail I, Frapy E, Dupuis M, Dieppedale J, Charbit A, Meibom KL. Identification of a novel small RNA modulating Francisella tularensis pathogenicity. PLoS One. 2012;7:e41999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 95. | Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, Sayood K, Dunman PM, Roop Ii RM. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol. 2012;85:345-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 96. | Schmidtke C, Findeiss S, Sharma CM, Kuhfuss J, Hoffmann S, Vogel J, Stadler PF, Bonas U. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 2012;40:2020-2031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 97. | Zeng Q, McNally RR, Sundin GW. Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J Bacteriol. 2013;195:1706-1717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 98. | Torres-Quesada O, Reinkensmeier J, Schlüter JP, Robledo M, Peregrina A, Giegerich R, Toro N, Becker A, Jiménez-Zurdo JI. Genome-wide profiling of Hfq-binding RNAs uncovers extensive post-transcriptional rewiring of major stress response and symbiotic regulons in Sinorhizobium meliloti. RNA Biol. 2014;11:563-579. [PubMed] |
| 99. | Xiao B, Li W, Guo G, Li B, Liu Z, Jia K, Guo Y, Mao X, Zou Q. Identification of small noncoding RNAs in Helicobacter pylori by a bioinformatics-based approach. Curr Microbiol. 2009;58:258-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 100. | Wang Q, Harshey RM. Rcs signalling-activated transcription of rcsA induces strong anti-sense transcription of upstream fliPQR flagellar genes from a weak intergenic promoter: regulatory roles for the anti-sense transcript in virulence and motility. Mol Microbiol. 2009;74:71-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 101. | Arnvig KB, Young DB. Identification of small RNAs in Mycobacterium tuberculosis. Mol Microbiol. 2009;73:397-408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 102. | Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, Perkins TT, Parkhill J, Dougan G. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 103. | DiChiara JM, Contreras-Martinez LM, Livny J, Smith D, McDonough KA, Belfort M. Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucleic Acids Res. 2010;38:4067-4078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 104. | Tsai CH, Baranowski C, Livny J, McDonough KA, Wade JT, Contreras LM. Identification of novel sRNAs in mycobacterial species. PLoS One. 2013;8:e79411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 105. | Pelly S, Bishai WR, Lamichhane G. A screen for non-coding RNA in Mycobacterium tuberculosis reveals a cAMP-responsive RNA that is expressed during infection. Gene. 2012;500:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 106. | Warner DF, Savvi S, Mizrahi V, Dawes SS. A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. J Bacteriol. 2007;189:3655-3659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 68] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 107. | Miotto P, Forti F, Ambrosi A, Pellin D, Veiga DF, Balazsi G, Gennaro ML, Di Serio C, Ghisotti D, Cirillo DM. Genome-wide discovery of small RNAs in Mycobacterium tuberculosis. PLoS One. 2012;7:e51950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 108. | Hartkoorn RC, Sala C, Uplekar S, Busso P, Rougemont J, Cole ST. Genome-wide definition of the SigF regulon in Mycobacterium tuberculosis. J Bacteriol. 2012;194:2001-2009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 109. | Houghton J, Cortes T, Schubert O, Rose G, Rodgers A, De Ste Croix M, Aebersold R, Young DB, Arnvig KB. A small RNA encoded in the Rv2660c locus of Mycobacterium tuberculosis is induced during starvation and infection. PLoS One. 2013;8:e80047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 110. | McGuire AM, Weiner B, Park ST, Wapinski I, Raman S, Dolganov G, Peterson M, Riley R, Zucker J, Abeel T. Comparative analysis of Mycobacterium and related Actinomycetes yields insight into the evolution of Mycobacterium tuberculosis pathogenesis. BMC Genomics. 2012;13:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 111. | Golby P, Nunez J, Witney A, Hinds J, Quail MA, Bentley S, Harris S, Smith N, Hewinson RG, Gordon SV. Genome-level analyses of Mycobacterium bovis lineages reveal the role of SNPs and antisense transcription in differential gene expression. BMC Genomics. 2013;14:710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 112. | Ignatov D, Malakho S, Majorov K, Skvortsov T, Apt A, Azhikina T. RNA-Seq analysis of Mycobacterium avium non-coding transcriptome. PLoS One. 2013;8:e74209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 113. | Li SK, Ng PK, Qin H, Lau JK, Lau JP, Tsui SK, Chan TF, Lau TC. Identification of small RNAs in Mycobacterium smegmatis using heterologous Hfq. RNA. 2013;19:74-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 114. | Pandey SP, Minesinger BK, Kumar J, Walker GC. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 2011;39:4691-4708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 115. | André G, Even S, Putzer H, Burguière P, Croux C, Danchin A, Martin-Verstraete I, Soutourina O. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 2008;36:5955-5969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 106] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 116. | Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell. 2004;14:647-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 137] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 117. | Stork M, Di Lorenzo M, Welch TJ, Crosa JH. Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J Bacteriol. 2007;189:3479-3488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 118. | Blomberg P, Wagner EG, Nordström K. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990;9:2331-2340. [PubMed] |
| 119. | Carpousis AJ, Luisi BF, McDowall KJ. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci. 2009;85:91-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 125] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 120. | Gerdes K, Nielsen A, Thorsted P, Wagner EG. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. J Mol Biol. 1992;226:637-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 96] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 121. | Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jäger JG, Hüttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435-6443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 349] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 122. | Saramago M, Bárria C, Dos Santos RF, Silva IJ, Pobre V, Domingues S, Andrade JM, Viegas SC, Arraiano CM. The role of RNases in the regulation of small RNAs. Curr Opin Microbiol. 2014;18:105-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 123. | Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5’ pyrophosphate removal. Mol Cell. 2007;27:79-90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 200] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 124. | Kaga N, Umitsuki G, Nagai K, Wachi M. RNase G-dependent degradation of the eno mRNA encoding a glycolysis enzyme enolase in Escherichia coli. Biosci Biotechnol Biochem. 2002;66:2216-2220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 125. | Umitsuki G, Wachi M, Takada A, Hikichi T, Nagai K. Involvement of RNase G in in vivo mRNA metabolism in Escherichia coli. Genes Cells. 2001;6:403-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 67] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 126. | Alifano P, Rivellini F, Piscitelli C, Arraiano CM, Bruni CB, Carlomagno MS. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 1994;8:3021-3031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 122] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 127. | Li Y, Altman S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc Natl Acad Sci USA. 2003;100:13213-13218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 128. | Iwamoto A, Lemire S, Yonesaki T. Post-transcriptional control of Crp-cAMP by RNase LS in Escherichia coli. Mol Microbiol. 2008;70:1570-1578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 129. | Perwez T, Kushner SR. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol Microbiol. 2006;60:723-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 130. | Schilling O, Rüggeberg S, Vogel A, Rittner N, Weichert S, Schmidt S, Doig S, Franz T, Benes V, Andrews SC. Characterization of an Escherichia coli elaC deletion mutant. Biochem Biophys Res Commun. 2004;320:1365-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 131. | Anupama K, Leela JK, Gowrishankar J. Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho-dependent transcription termination. Mol Microbiol. 2011;82:1330-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 132. | Even S, Pellegrini O, Zig L, Labas V, Vinh J, Bréchemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 2005;33:2141-2152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 252] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 133. | Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28:3523-3533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 203] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 134. | Herrmann B, Stolt P, Abdeldaim G, Rubin CJ, Kirsebom LA, Thollesson M. Differentiation and phylogenetic relationships in Mycobacterium spp with special reference to the RNase P RNA gene rnpB. Curr Microbiol. 2014;69:634-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 135. | Taverniti V, Forti F, Ghisotti D, Putzer H. Mycobacterium smegmatis RNase J is a 5’-3’ exo-/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Mol Microbiol. 2011;82:1260-1276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 136. | Abendroth J, Ollodart A, Andrews ES, Myler PJ, Staker BL, Edwards TE, Arcus VL, Grundner C. Mycobacterium tuberculosis Rv2179c protein establishes a new exoribonuclease family with broad phylogenetic distribution. J Biol Chem. 2014;289:2139-2147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 137. | Murdeshwar MS, Chatterji D. MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J Bacteriol. 2012;194:4003-4014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 138. | Kovacs L, Csanadi A, Megyeri K, Kaberdin VR, Miczak A. Mycobacterial RNase E-associated proteins. Microbiol Immunol. 2005;49:1003-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 139. | Watkins HA, Baker EN. Structural and functional characterization of an RNase HI domain from the bifunctional protein Rv2228c from Mycobacterium tuberculosis. J Bacteriol. 2010;192:2878-2886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 140. | Csanadi A, Faludi I, Miczak A. MSMEG_4626 ribonuclease from Mycobacterium smegmatis. Mol Biol Rep. 2009;36:2341-2344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 141. | Zeller ME, Csanadi A, Miczak A, Rose T, Bizebard T, Kaberdin VR. Quaternary structure and biochemical properties of mycobacterial RNase E/G. Biochem J. 2007;403:207-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 142. | Akey DL, Berger JM. Structure of the nuclease domain of ribonuclease III from M. tuberculosis at 2.1 A. Protein Sci. 2005;14:2744-2750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 143. | Kittle JD, Simons RW, Lee J, Kleckner N. Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5’ end of the target RNA and a loop in the anti-sense RNA. J Mol Biol. 1989;210:561-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 144. | Tomizawa J. Control of ColE1 plasmid replication. Interaction of Rom protein with an unstable complex formed by RNA I and RNA II. J Mol Biol. 1990;212:695-708. [PubMed] |
| 145. | Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984;38:861-870. [PubMed] |
| 146. | Franch T, Gerdes K. U-turns and regulatory RNAs. Curr Opin Microbiol. 2000;3:159-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 52] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 147. | Nakashima N, Tamura T, Good L. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli. Nucleic Acids Res. 2006;34:e138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 98] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 148. | Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. Bactericidal antisense effects of peptide-PNA conjugates. Nat Biotechnol. 2001;19:360-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 311] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 149. | Nikravesh A, Dryselius R, Faridani OR, Goh S, Sadeghizadeh M, Behmanesh M, Ganyu A, Klok EJ, Zain R, Good L. Antisense PNA accumulates in Escherichia coli and mediates a long post-antibiotic effect. Mol Ther. 2007;15:1537-1542. [PubMed] |
| 150. | Rasmussen LC, Sperling-Petersen HU, Mortensen KK. Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb Cell Fact. 2007;6:24. [PubMed] |
| 151. | Cao M, Ren H, Zhao P, Pan W, Zhao L, Qi Z. Small interfering RNA-mediated inhibition of hepatitis G virus gene expression in human hepatoma cell Huh-7. Sci China C Life Sci. 2005;48:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 152. | Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, Chen J. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA. 2003;100:2718-2723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 405] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 153. | Lim T, Yuan J, Zhang HM, Sall A, Liu Z, Su Y, Yang D. Antisense DNA and RNA agents against picornaviruses. Front Biosci. 2008;13:4707-4725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 154. | Zhang JM, Guo J, Tu X, Shi ZH, Hao JJ, Ke YH, Guan JF, He JJ. [Protective effect of Huaxia shallot preparation on human umbilical vein endothelial cell injury induced by oxidized low density lipoprotein and its mechanism]. Zhongxiyi Jiehe Xuebao. 2007;5:675-680. [PubMed] |
| 155. | Mellbye BL, Weller DD, Hassinger JN, Reeves MD, Lovejoy CE, Iversen PL, Geller BL. Cationic phosphorodiamidate morpholino oligomers efficiently prevent growth of Escherichia coli in vitro and in vivo. J Antimicrob Chemother. 2010;65:98-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 156. | Tilley LD, Mellbye BL, Puckett SE, Iversen PL, Geller BL. Antisense peptide-phosphorodiamidate morpholino oligomer conjugate: dose-response in mice infected with Escherichia coli. J Antimicrob Chemother. 2007;59:66-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 157. | Mitev GM, Mellbye BL, Iversen PL, Geller BL. Inhibition of intracellular growth of Salmonella enterica serovar Typhimurium in tissue culture by antisense peptide-phosphorodiamidate morpholino oligomer. Antimicrob Agents Chemother. 2009;53:3700-3704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 158. | Nekhotiaeva N, Awasthi SK, Nielsen PE, Good L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol Ther. 2004;10:652-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 124] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 159. | George R, Nugent K, Bolus N, Garner J, Pickering J, Glaze A, Heard E, Waugh J, Unlap M. A Short Interfering RNA Molecular Beacon for the Attenuation of Mycobacterial Infection. AJBB. 2014;10:40-49. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 160. | Harth G, Horwitz MA, Tabatadze D, Zamecnik PC. Targeting the Mycobacterium tuberculosis 30/32-kDa mycolyl transferase complex as a therapeutic strategy against tuberculosis: Proof of principle by using antisense technology. Proc Natl Acad Sci USA. 2002;99:15614-15619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 161. | Borden JR, Jones SW, Indurthi D, Chen Y, Papoutsakis ET. A genomic-library based discovery of a novel, possibly synthetic, acid-tolerance mechanism in Clostridium acetobutylicum involving non-coding RNAs and ribosomal RNA processing. Metab Eng. 2010;12:268-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 162. | Goh S, Boberek JM, Nakashima N, Stach J, Good L. Concurrent growth rate and transcript analyses reveal essential gene stringency in Escherichia coli. PLoS One. 2009;4:e6061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 163. | Ji Y, Zhang B, Van SF, Horn P, Woodnutt G, Burnham MK, Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266-2269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 333] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 164. | Tummala SB, Junne SG, Paredes CJ, Papoutsakis ET. Transcriptional analysis of product-concentration driven changes in cellular programs of recombinant Clostridium acetobutylicumstrains. Biotechnol Bioeng. 2003;84:842-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 165. | Perret S, Maamar H, Bélaich JP, Tardif C. Use of antisense RNA to modify the composition of cellulosomes produced by Clostridium cellulolyticum. Mol Microbiol. 2004;51:599-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 166. | Ji Y, Marra A, Rosenberg M, Woodnutt G. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J Bacteriol. 1999;181:6585-6590. [PubMed] |
| 167. | Morrissey JA, Cockayne A, Hill PJ, Williams P. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect Immun. 2000;68:6281-6288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 168. | Parish CA, de la Cruz M, Smith SK, Zink D, Baxter J, Tucker-Samaras S, Collado J, Platas G, Bills G, Díez MT. Antisense-guided isolation and structure elucidation of pannomycin, a substituted cis-decalin from Geomyces pannorum. J Nat Prod. 2009;72:59-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 169. | Zhang C, Ondeyka JG, Zink DL, Basilio A, Vicente F, Collado J, Platas G, Huber J, Dorso K, Motyl M. Isolation, structure and antibacterial activity of pleosporone from a pleosporalean ascomycete discovered by using antisense strategy. Bioorg Med Chem. 2009;17:2162-2166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 170. | Gruegelsiepe H, Brandt O, Hartmann RK. Antisense inhibition of RNase P: mechanistic aspects and application to live bacteria. J Biol Chem. 2006;281:30613-30620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 171. | Ji Y, Yin D, Fox B, Holmes DJ, Payne D, Rosenberg M. Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol Lett. 2004;231:177-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 172. | Jeon B, Zhang Q. Sensitization of Campylobacter jejuni to fluoroquinolone and macrolide antibiotics by antisense inhibition of the CmeABC multidrug efflux transporter. J Antimicrob Chemother. 2009;63:946-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 173. | Kedar GC, Brown-Driver V, Reyes DR, Hilgers MT, Stidham MA, Shaw KJ, Finn J, Haselbeck RJ. Evaluation of the metS and murB loci for antibiotic discovery using targeted antisense RNA expression analysis in Bacillus anthracis. Antimicrob Agents Chemother. 2007;51:1708-1718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 174. | Hong H, Zhang Y, Cai W. In vivo imaging of RNA interference. J Nucl Med. 2010;51:169-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 175. | Kim Y, Sohn D, Tan W. Molecular beacons in biomedical detection and clinical diagnosis. Int J Clin Exp Pathol. 2008;1:105-116. [PubMed] |
| 176. | Ilieva M, Della Vedova P, Hansen O, Dufva M. Tracking neuronal marker expression inside living differentiating cells using molecular beacons. Front Cell Neurosci. 2013;7:266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 177. | Chang E, Zhu MQ, Drezek R. Novel siRNA-based molecular beacons for dual imaging and therapy. Biotechnol J. 2007;2:422-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 178. | Rhee WJ, Santangelo PJ, Jo H, Bao G. Target accessibility and signal specificity in live-cell detection of BMP-4 mRNA using molecular beacons. Nucleic Acids Res. 2008;36:e30. [PubMed] |
| 179. | Zhou C, Mao Y, Sugimoto Y, Zhang Y, Kanthamneni N, Yu B, Brueggemeier RW, Lee LJ, Lee RJ. SPANosomes as delivery vehicles for small interfering RNA (siRNA). Mol Pharm. 2012;9:201-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 180. | George R, Bolus N, Garner J, Nugent K, Unlap MT. A Short Interfering RNA (siRNA) Molecular Beacon for the Detection of Mycobacterial Infection. J Biotechnol Biomater. 2012;2:1000147. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 181. | Haile Y, Caugant DA, Bjune G, Wiker HG. Mycobacterium tuberculosis mammalian cell entry operon (mce) homologs in Mycobacterium other than tuberculosis (MOTT). FEMS Immunol Med Microbiol. 2002;33:125-132. [PubMed] |
| 182. | Anes E, Peyron P, Staali L, Jordao L, Gutierrez MG, Kress H, Hagedorn M, Maridonneau-Parini I, Skinner MA, Wildeman AG. Dynamic life and death interactions between Mycobacterium smegmatis and J774 macrophages. Cell Microbiol. 2006;8:939-960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 92] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 183. | Xu G, Li Y, Yang J, Zhou X, Yin X, Liu M, Zhao D. Effect of recombinant Mce4A protein of Mycobacterium bovis on expression of TNF-alpha, iNOS, IL-6, and IL-12 in bovine alveolar macrophages. Mol Cell Biochem. 2007;302:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 184. | Nyce J. Respirable antisense oligonucleotides: a new, third drug class targeting respiratory disease. Curr Opin Allergy Clin Immunol. 2002;2:533-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 185. | Mönkkönen J, Urtti A. Lipid fusion in oligonucleotide and gene delivery with cationic lipids. Adv Drug Deliv Rev. 1998;34:37-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 63] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 186. | Nyce JW, Metzger WJ. DNA antisense therapy for asthma in an animal model. Nature. 1997;385:721-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 171] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Bugaj AM, Hattori N S- Editor: Ji FF L- Editor: A E- Editor: Jiao XK