Published online Sep 9, 2025. doi: 10.5492/wjccm.v14.i3.106496
Revised: April 11, 2025
Accepted: May 7, 2025
Published online: September 9, 2025
Processing time: 141 Days and 10.4 Hours
Nosocomial fever of unknown origin (nFUO) is a frequent and challenging diagnostic entity, encompassing diverse infectious and non-infectious etiologies. Timely identification is crucial, yet evidence on the diagnostic accuracy of com
To evaluate the diagnostic utility of sepsis tools and biomarkers in identifying infectious causes of nFUO.
This prospective observational study included patients admitted to the Acute Care Emergency Medicine Unit, Postgraduate Institute of Medical Education and Research, Chandigarh, India (July 2023 to December 2024). nFUO was defined by Durack and Street criteria. Diagnostic performance of sepsis screening tools (systemic inflammatory response syndrome, Sequential Organ Failure Asse
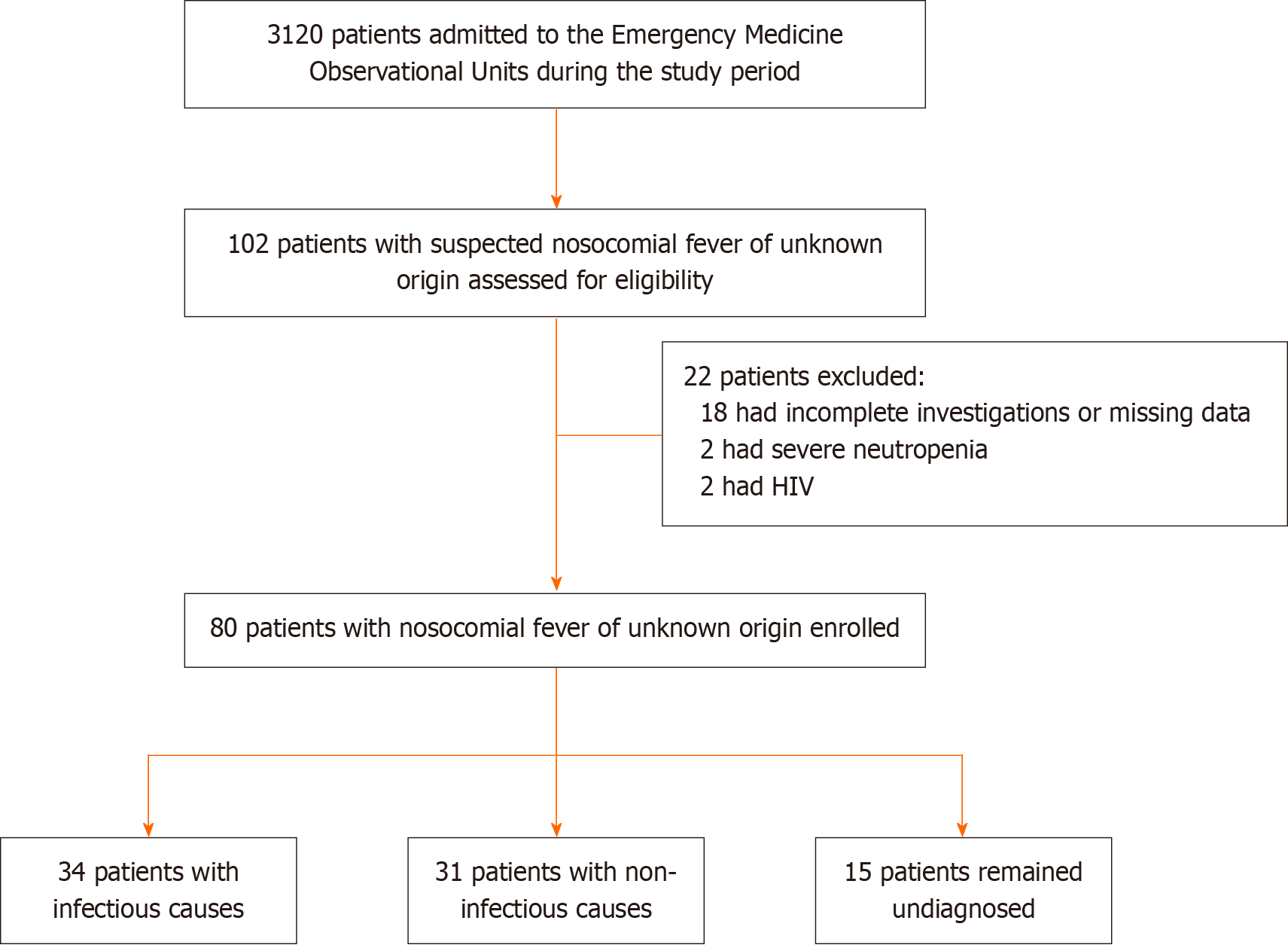
Of 80 cases (mean age 42.9 ± 16.5 years; 80% male), 42.5% had infectious causes, 38.7% non-infectious, and 18.8% remained undiagnosed. Pneumonia (26.2%) and bloodstream infections (11.2%) were the most common infectious etiologies, while central fever and thrombophlebitis (each 7.5%) were predominant among non-infectious causes. Sepsis tools showed poor diagnostic accuracy, with area under the receiver operating characteristic curve (AUC) values close to 0.5. PCT demonstrated modest performance (AUC = 0.61; optimal cut-off: 0.85 μg/L), while CRP was paradoxically higher in non-infectious cases (AUC = 0.45). Overall mortality was 20% and was highest among undiagnosed patients (33.3%). Fever duration and hospitalization length were significantly greater in infectious cases.
Sepsis tools, PCT, and CRP have limited utility in identifying infectious causes of nFUO in critically ill adults and should not solely guide initial decision-making.
Core Tip: Nosocomial fever of unknown origin is a frequent and complex challenge in critically ill patients, requiring prompt differentiation between infectious and non-infectious causes. This study evaluates the diagnostic performance of sepsis screening tools, procalcitonin, and C-reactive protein at fever onset. Findings reveal limited utility of all sepsis tools. Procalcitonin shows modest accuracy, while C-reactive protein is unreliable. Given the associated high mortality, this study emphasizes the importance of a structured, systematic evaluation over empirical antibiotic use, and highlights the need for advanced diagnostic modalities to improve infection detection.
- Citation: Saini S, Pahil S, Mohindra R, Sachdeva N, Sharma N, Pannu AK. Diagnostic utility of sepsis screening tools, procalcitonin, and C-reactive protein in nosocomial fever of unknown origin. World J Crit Care Med 2025; 14(3): 106496
- URL: https://www.wjgnet.com/2220-3141/full/v14/i3/106496.htm
- DOI: https://dx.doi.org/10.5492/wjccm.v14.i3.106496
Prolonged, unexplained new-onset fever in hospitalized patients, commonly referred to as nosocomial fever of unknown origin (nFUO), is a frequent and challenging phenomenon[1,2]. Despite its clinical significance, nFUO remains under-researched, with limited studies exploring its spectrum, diagnostic and management strategies, and outcomes since Durack and Street (1991) first described it[1-5]. Existing literature highlights its multifactorial etiology and diagnostic challenges, underscoring the need for a systematic approach to the evaluation and management of nFUO[2-5].
Causes of nFUO can be broadly categorized into infectious and non-infectious origins. Infectious causes include pneumonia, such as ventilator-associated pneumonia (VAP) or hospital-acquired pneumonia (HAP), bloodstream infections such as catheter-related bloodstream infection, fungemia, catheter-associated urinary tract infection, Clostridioides difficile infection, and pressure ulcers[2-5]. Non-infectious etiologies encompass venous thromboembolism such as deep vein thrombosis and pulmonary embolism, acute infarctions, including myocardial infarction and stroke, cerebral hemorrhages, acalculous cholecystitis, drug fever, transfusion-related reactions, hemolysis, and bleeding[2-5]. Identifying the underlying cause of nFUO is critical, as it often leads to prolonged hospital stays, increased morbidity and mortality, and delayed or inappropriate treatments.
Early initiation of empirical antibiotic therapy for new-onset fever in hospitalized patients, particularly in critical care settings, is justified to improve infection-related outcomes[6]. However, when initial cultures are sterile and an infection is not apparent, this approach carries the risk of unnecessary antibiotic use, contributing to the global issue of antimicrobial resistance[7]. Unlike classical FUO where empirical antimicrobial therapy is often discouraged, there are no such definitive recommendations for nFUO, likely due to the predominance of bacterial etiologies[2,8]. This raises a clinically relevant question: Can we reliably differentiate between infectious and non-infectious causes of nFUO at its onset?
Sepsis screening tools, such as systemic inflammatory response syndrome (SIRS) criteria, Sequential Organ Failure Assessment (SOFA) score, quick SOFA (qSOFA) score, National Early Warning Score (NEWS), quick NEWS (qNEWS), and Modified Early Warning Score (MEWS), along with sepsis biomarkers with rapid turnaround times, like procalcitonin (PCT) and C-reactive protein (CRP), are commonly used for early sepsis diagnosis and to guide antimicrobial therapy in acute care settings[9,10]. While these tools hold promise for evaluating new fever in critically ill patients, the Society of Critical Care Medicine and Infectious Diseases Society of America (SCCM/IDSA) 2023 guidelines do not discuss the role of sepsis screening tools[6]. Although the guidelines discuss the utility of PCT and CRP, their recommendations focus on distinguishing infectious from non-infectious causes of new-onset fever rather than specifically addressing nFUO[6].
Thus, the diagnostic accuracy and clinical utility of sepsis screening tools and biomarkers in differentiating infectious from non-infectious causes of nFUO remain inadequately studied. Addressing this critical knowledge gap is essential to optimize the management of nFUO, minimize unnecessary antibiotic use, and improve patient outcomes. This study aims to evaluate the utility of various sepsis screening tools, PCT, and CRP at fever onset in distinguishing between infectious and non-infectious causes of nFUO in acute care settings.
This prospective observational cohort study was conducted in the Acute Care and Emergency Medicine (ACEM) Unit of the Department of Internal Medicine at the Postgraduate Institute of Medical Education and Research, Chandigarh, India, from July 2023 to December 2024. A formal sample size calculation was not performed, given the exploratory nature of this prospective diagnostic study and the absence of prior data to estimate a precise effect size. Instead, a convenience sampling strategy was used, enrolling all consecutive eligible patients during the predefined 18-month study period. This pragmatic approach reflects real-world clinical practice and was intended to generate preliminary data in an underexplored area. The ACEM unit manages approximately 120-150 daily admissions, encompassing the Emergency Rooms and Observation Units, including a High Dependency Unit[11].
Inclusion criteria including: (1) Adults (≥ 18 years) admitted to the ACEM unit; and (2) Patients fulfilling the diagnostic definition of nFUO based on Durack and Street criteria (1991): Fever ≥ 101°F on multiple occasions after ≥ 48 hours of hospitalization; infection not present or incubating at the time of admission; and no definitive diagnosis despite 3 days of inpatient investigations, including 24-hour incubation of microbiological cultures[1].
Exclusion criteria including: (1) Known immunocompromised states, including persons with human immunodeficiency virus, severe neutropenia (absolute neutrophil count < 500/μL), or history of solid organ or hematopoietic stem cell transplantation; (2) Fever present at the time of admission or developing within 48 hours of hospitalization; and (3) Incomplete data or withdrawal of consent.
The initial evaluation of new-onset fever was conducted following the SCCM/IDSA 2023 guidelines for critically ill adult patients[6]. Clinical evaluation included an assessment of new-onset symptoms or signs, a vital examination, and a detailed drug history. Baseline sepsis screening tools, including SIRS, qSOFA, SOFA, NEWS, qNEWS, and MEWS, were assessed at fever onset[9,10]. Chronic comorbidities were evaluated using the validated Charlson comorbidity index and the age-adjusted Charlson comorbidity index, with higher scores indicating a greater burden of comorbidities and increased risk of mortality[12,13]. Acute medical disorders leading to patient admissions in the ACEM unit were defined and categorized according to the International Classification of Diseases and Related Health Problems, 11th Revision[14].
Laboratory investigations at fever onset included complete blood count, biochemistry panel (serum electrolytes, renal function tests, bilirubin, liver enzymes, total protein, and albumin), sepsis biomarkers (serum PCT and CRP), appropriate cultures, and chest radiography. PCT levels were measured using electrochemiluminescence immunoassay (range: 0.02-100.00 μg/L; reference < 0.5 μg/L), while CRP levels were measured via particle-enhanced immunoturbidimetric assay (range 0.6-350.0 mg/L; reference < 5 mg/L).
Blood cultures, preferably two sets, were obtained from all patients before initiating antibiotics. For patients already on antibiotics, cultures were collected immediately before the next dose. In patients with central venous catheters (CVCs), simultaneous cultures were obtained from both the CVC and a peripheral venipuncture site. Blood cultures were processed using the BACTEC™ system, an automated continuous monitoring system that detects microbial growth by measuring increases in carbon dioxide concentration via fluorescence. Antibiotic susceptibility testing was performed using the Kirby-Bauer disc diffusion method and the Vitek 2 system (BioMérieux), following Clinical and Laboratory Standards Institute guidelines[15].
Further laboratory tests and imaging studies were guided by potentially diagnostic clues, defined as any clinical or laboratory finding suggesting a potential etiology for fever[2,6]. For example, urine cultures were performed in patients with gross or microscopic pyuria or when urinary tract infection was clinically suspected. Abdominal ultrasonography was conducted when an abdominal source of fever was suspected. Advanced imaging modalities such as computed tomography (CT), magnetic resonance imaging (MRI), or 18F-fluorodeoxyglucose positron emission tomography (FDG-PET) were considered when initial diagnostic evaluations failed to establish a cause, provided patient transport was deemed safe[6].
Patient management adhered to standard guidelines, including the Surviving Sepsis Campaign (2021) and SCCM/IDSA (2023) recommendations[6,9]. Intensive organ support, defined as invasive mechanical ventilation, vasopressors to maintain a mean arterial pressure of ≥ 70 mmHg, and renal replacement therapy (e.g., hemodialysis), was provided as required. Patients were followed until discharge or death, and outcomes such as mortality, fever resolution and its duration, and length of hospital stay were recorded.
The etiology of nFUO was classified into three categories: (1) Infectious disorders: Diagnosed based on the Centers for Disease Control and Prevention and National Healthcare Safety Network surveillance definitions for healthcare-associated infections[16]; (2) Non-infectious disorders: Conditions such as central fever, acute pancreatitis, aspiration pneumonitis (chemical) or pneumonitis without infections, venous thromboembolism, and thrombophlebitis were defined using standard guidelines and criteria[17-19]. Central fever was diagnosed in patients with negative blood and body fluid cultures, absence of infiltrates on chest radiography, and fever onset within 72 hours of admission, alongside a primary diagnosis of neurological conditions such as subarachnoid hemorrhage, intraventricular hemorrhage, extensive infarction with significant space-occupying brain edema, or tumor[17]. Suspected drug fever was evaluated using the Naranjo Adverse Drug Reaction Probability Scale to establish causality[20]; and (3) Undiagnosed: Cases in which the etiology of nFUO could not be determined despite thorough and appropriate investigations during the hospital stay.
Statistical analysis was performed using SPSS Statistics software, version 25.0 for Mac. Categorical variables were expressed as numbers and percentages, while continuous variables were presented as mean with standard deviation or median with interquartile range (IQR), depending on their distribution. The Shapiro-Wilk test was used to assess the normality of continuous data. Comparisons between infectious and non-infectious FUO groups were performed using the χ2 test for categorical variables, with Fisher’s exact test applied when expected frequencies were small. For continuous variables, analysis of variance was used for normally distributed data, and the Mann-Whitney U test was used for non-normally distributed data.
Receiver operating characteristic (ROC) curve analysis was conducted to evaluate the diagnostic accuracy of sepsis screening tools and biomarkers in differentiating between infectious and non-infectious nFUO. The area under the ROC curve (AUC) was calculated to measure discriminatory performance, with values closer to 1.0 indicating excellent discrimination. Optimal thresholds for sepsis biomarkers were determined using Youden’s J statistic to maximize sensitivity and specificity. All statistical tests were two-tailed, and a P value of < 0.05 was considered statistically significant.
A total of 80 patients were enrolled, with a mean age of 42.9 ± 16.5 years (range: 18-75 years) and a male predominance (80%). A flowchart of patient enrollment and classification is shown in Figure 1. Infectious causes accounted for 42.5% of cases, non-infectious for 38.7% of cases, and 18.8% of cases remained undiagnosed (Table 1).
| Etiologies | n (%) |
| Infectious disorders | 34 (42.5) |
| Pneumonia | 21 (26.2) |
| Ventilator-associated pneumonia1 | 17 |
| Hospital-acquired pneumonia2 | 4 |
| Bloodstream infections | 9 (11.2) |
| Bacteraemia with unknown primary site3 | 6 |
| Catheter-related blood stream infection4 | 3 |
| Intra-abdominal Infections | 3 (3.75) |
| Clostridioides difficile diarrhea | 1 |
| Peritonitis | 1 |
| Gall bladder perforation with peritonitis and intra-abdominal collections | 1 |
| Catheter-associated urinary tract infection5 | 1 (1.3) |
| Multi-site infections | 2 (2.5) |
| Ventilator-associated pneumonia and pressure ulcer6 | 1 |
| Ventilator-associated pneumonia7 and catheter-related blood stream infection8 | 1 |
| Non-infectious disorders | 31 (38.7) |
| Central fever | 6 (7.5) |
| Thrombophlebitis | 6 (7.5) |
| Pneumonitis without infection | 4 (5) |
| Acute pancreatitis | 3 (3.8) |
| Rhabdomyolysis | 3 (3.8) |
| Venous thromboembolism | 2 (2.5) |
| Drug fever | 2 (2.5) |
| Alcohol withdrawal syndrome | 1 (1.3) |
| Central pontine demyelination | 1 (1.3) |
| Transfusion reaction | 1 (1.3) |
| Acute hemolysis | 1 (1.3) |
| Atelectasis | 1 (1.3) |
| Undiagnosed | 15 (18.8) |
Pre-existing comorbidities were present in 60% of patients. Neurological (40%) and toxicological (23.7%) emergencies were the most frequent admitting diagnoses. There were no significant differences in baseline comorbidities, acute medical illnesses, vital signs, the requirement for intensive organ support, or the presence of various in-situ catheters and tubes at the time of fever onset between the infectious and non-infectious groups (Table 2). The median time to fever onset post-hospitalization was 5 days (IQR: 3-8; range: 3-25). Baseline laboratory characteristics, including leukocytosis, neutrophil count, neutrophil-to-lymphocyte ratio, and plasma albumin, were comparable between groups (Table 3).
| Parameter | Total (n = 80) | Infectious (n = 34) | Non-infectious (n = 31) | P value |
| Age (years), mean ± SD | 42.9 ± 16.5 | 39.2 ± 17.4 | 45.8 ± 14.1 | 0.098 |
| Male gender | 64 (80) | 26 (76.5) | 26 (83.9) | 0.456 |
| Pre-existing comorbidities | ||||
| Any | 48 (60) | 21 (61.8) | 20 (64.5) | 0.818 |
| Diabetes | 19 (23.8) | 7 (20.6) | 8 (25.8) | 0.618 |
| Hypertension | 14 (17.5) | 7 (20.6) | 4 (12.9) | 0.409 |
| Chronic kidney disease | 8 (10) | 2 (5.9) | 4 (12.9) | 0.413 |
| Epilepsy | 8 (10) | 5 (14.7) | 3 (9.7) | 0.711 |
| Coronary artery disease or heart failure | 7 (8.8) | 3 (8.8) | 4 (12.9) | 0.701 |
| Miscellaneous1 | 11 (13.8) | 5 (14.7) | 5 (16.1) | - |
| Charlson comorbidity index, median (IQR) | 0 (0-2) | 0 (0-1.25) | 0 (0-2) | 0.719 |
| Age-adjusted Charlson comorbidity index, median (IQR) | 0 (0-2) | 0 (0-2) | 1 (0-2) | 0.389 |
| Substance use disorders2 | 20 (25) | 5 (14.7) | 9 (29) | 0.161 |
| Medical emergencies | ||||
| Neurological disorders3 | 32 (40) | 14 (41.2) | 16 (51.6) | 0.399 |
| Toxicological disorders4 | 19 (23.7) | 6 (17.6) | 9 (29) | 0.277 |
| Gastro-intestinal disorders5 | 7 (8.75) | 3 (8.8) | 1 (3.2) | 0.615 |
| Renal disorders6 | 6 (7.5) | 2 (5.9) | 2 (6.5) | 1.000 |
| Miscellaneous7 | 16 (20) | 9 (26.5) | 3 (9.7) | - |
| Parameter | Total (n = 80) | Infectious (n = 34) | Non-infectious (n = 31) | P value |
| Time to fever onset post-hospitalization (days), median (IQR) | 5 (3-8) | 5 (3.75-9) | 5 (3-7) | 0.229 |
| Temperature (F), mean ± SD | 101.6 ± 0.7 | 101.7 ± 0.8 | 101.5 ± 0.7 | 0.714 |
| Pulse rate (per minute), mean ± SD | 104.2 ± 20.6 | 109.0 ± 23.4 | 100.0 ± 16.0 | 0.079 |
| Tachycardia, n (%) | 36 (45) | 18 (52.9) | 12 (38.7) | 0.250 |
| Systolic blood pressure (mmHg), mean ± SD | 127.9 ± 21.9 | 128.8 ± 23.6 | 126.3 ± 21.2 | 0.655 |
| Diastolic blood pressure (mmHg), mean ± SD | 76.8 ± 12.9 | 79.5 ± 14.5 | 74.4 ± 11.2 | 0.118 |
| Mean arterial pressure (mmHg), mean ± SD | 93.8 ± 14.6 | 95.9 ± 16.3 | 91.67 ± 13.7 | 0.262 |
| Glasgow coma scale, median (IQR) | 10 (8-11) | 10 (8-14.3) | 9 (7.0-11.0) | 0.185 |
| Intensive organ support requirement | ||||
| Mechanical ventilation, n (%) | 64 (80) | 28 (82.4) | 24 (77.4) | 0.619 |
| Vasopressor therapy, n (%) | 11 (13.8) | 5 (14.7) | 4 (12.9) | 1.000 |
| Hemodialysis, n (%) | 8 (10) | 5 (14.7) | 0 (0) | 0.054 |
| In-situ catheters or tubes, n (%) | ||||
| Urinary catheters | 72 (90) | 31 (91.2) | 29 (93.5) | 1.000 |
| Endotracheal tubes | 64 (80) | 28 (82.4) | 24 (77.4) | 0.619 |
| Nasogastric tubes | 64 (80) | 29 (85.3) | 24 (77.4) | 0.414 |
| Central lines | 25 (31.3) | 9 (26.5) | 5 (16.1) | 0.311 |
| Non-tunnelled dialysis catheters | 7 (8.8) | 4 (11.8) | 0 (0) | 0.115 |
| Percutaneous catheters | 6 (7.5) | 3 (8.8) | 1 (3.2) | 0.614 |
| Laboratory investigations | ||||
| Hemoglobin (g/L), mean ± SD | 110.7 ± 27.4 | 109.0 ± 28.2 | 109.6 ± 28.1 | 0.933 |
| Platelet count (per μL), median (IQR) | 165500 (109250-230250) | 168000 (125500-237250) | 198000 (110000-254000) | 0.590 |
| Total leucocyte count (per μL), median (IQR) | 12640 (10005-18842) | 12490 (9500-19165) | 14420 (1.280-23400) | 0.123 |
| Leukocytosis, n (%) | 54 (67.5) | 22 (64.7) | 23 (74.2) | 0.408 |
| Neutrophils (%), mean ± SD | 82.7 ± 7.0 | 82.2 ± 6.4 | 84.3 ± 6.2 | 0.199 |
| Lymphocytes (%), mean ± SD | 8.2 ± 4.4 | 8.2 ± 4.1 | 7.8 ± 4.4 | 0.610 |
| Neutrophil to lymphocyte ratio, median (IQR) | 10.6 (7.2-20.7) | 10.5 (6.5-18.7) | 12.0 (7.4-23.7) | 0.470 |
| Sodium (mmol/L), mean ± SD | 141.0 ± 8.2 | 140.2 ± 7.2 | 142.0 ± 8.7 | 0.368 |
| Blood urea (mmol/L), median (IQR) | 8.7 (5.1-15.4) | 6.8 (4.4-11.2) | 9.2 (5.0-16.3) | 0.164 |
| Creatinine (mol/L), median (IQR) | 91.1 (61.9-209.7) | 97.2 (57.5-296.1) | 92.9 (61.9-210.4) | 0.478 |
| Bilirubin (mol/L), median (IQR) | 13.5 (6.8-23.6) | 12.6 (8.6-27.4) | 13.5 (6.7-20.0) | 0.454 |
| Aspartate transaminase (U/L), median (IQR) | 45.7 (25.9-87.8) | 44.5 (28.3-85.9) | 49.5 (23.0-102.0) | 0.984 |
| Alanine transaminase (U/L), median (IQR) | 32.1 (17.5-69.3) | 35.1 (19.8-57.0) | 29.2 (16.2-74.0) | 0.778 |
| Alkaline phosphatase (U/L), median (IQR) | 101 (75.3-132.8) | 92.5 (71-123.3) | 103 (75.5-133.5) | 0.711 |
| Total protein (g/L), mean ± SD | 63.9 ± 10.3 | 62.2 ± 9.7 | 64.4 ± 11.5 | 0.418 |
| Albumin (g/L), mean ± SD | 31.6 ± 5.8 | 31.2 ± 5.7 | 31.4 ± 5.6 | 0.863 |
The initial microbiological workup for new-onset fever included comprehensive hematological, biochemical, microbiological, and radiological investigations but none yielded potential diagnostic clues. Blood cultures were obtained for all patients (n = 80). Additional microbiological testing included microscopy and cultures of urine (n = 34), endotracheal aspirates (n = 27), sputum (n = 2), cerebrospinal fluid (n = 10), pleural fluid (n = 1), and wound swabs (n = 1). Radiological imaging comprised chest radiography (n = 80), head CT (n = 18), and abdominal ultrasonography (n = 15). Empirical antibiotic therapy was initiated at fever onset in all patients.
Following nFUO diagnosis, additional microbiological investigations were performed as indicated. These included repeat blood cultures (n = 52) and microscopy and cultures of endotracheal aspirates (n = 44), sputum (n = 5), urine (n = 26), cerebrospinal fluid (n = 4), wound swabs (n = 3), pleural fluid (n = 3), ascitic fluid (n = 2), peripancreatic collections (n = 2), CVC tip (n = 1), knee synovial fluid (n = 1), and stool samples (n = 1). Microbiological diagnoses were established from 23 respiratory cultures (21 endotracheal aspirates, 2 sputum), 9 blood cultures, 1 urine culture, 1 CVC tip culture, and 1 stool sample positive for C. difficile toxin.
Radiological investigations for nFUO included repeat chest radiography (n = 77), CT chest/abdomen (n = 17), MRI brain (n = 14), CT head (n = 12), abdominal ultrasonography (n = 9), and compression ultrasonography of lower limbs (n = 4). Imaging identified potentially diagnostic clues in the following cases: Chest radiography abnormalities (n = 28), including pneumonia (n = 23), pneumonitis without infection (n = 4), and atelectasis (n = 1); CT head findings suggestive of central fever (n = 6); CT abdomen findings consistent with peritonitis (n = 2) and pancreatitis (n = 1); CT chest showing pulmonary embolism (n = 1); MRI brain indicating central pontine demyelination (n = 1); and compression ultrasonography confirming deep vein thrombosis (n = 1).
Central fever was diagnosed in patients with intraventricular hemorrhage (n = 4) or large ischemic strokes (n = 2) after excluding infections. Pneumonitis or inflammatory lung injury (n = 4) was considered in cases with a history or risk of aspiration, supported by characteristic imaging findings and negative repeated respiratory and blood cultures. Thrombophlebitis (n = 6) was identified through careful clinical examinations, detecting subtle signs missed during the initial fever evaluation, and confirmed after exclusion of other etiologies.
Among the infectious disorders, all cases of pneumonia (VAP/HAP), bloodstream infections, and catheter-associated urinary tract infection were confirmed by culture. One case of C. difficile diarrhea was diagnosed based on C. difficile toxin assay. Peritonitis and gall bladder perforation with intra-abdominal collections were diagnosed based on CT abdomen findings in conjunction with neutrocytic ascites.
A majority of the cultured isolates (n = 29, 82.86%) were either extensively drug-resistant or multi-drug-resistant. All Acinetobacter baumannii strains (n = 21) were extensively drug-resistant, 14 strains were sensitive only to colistin and minocycline, five to colistin, and two to minocycline. Other multi-drug-resistant organisms included Klebsiella pneumoniae (n = 2), Pseudomonas aeruginosa (n = 1), Providencia stuartii (n = 1), Enterococcus faecium (n = 3), and Staphylococcus aureus (n = 1).
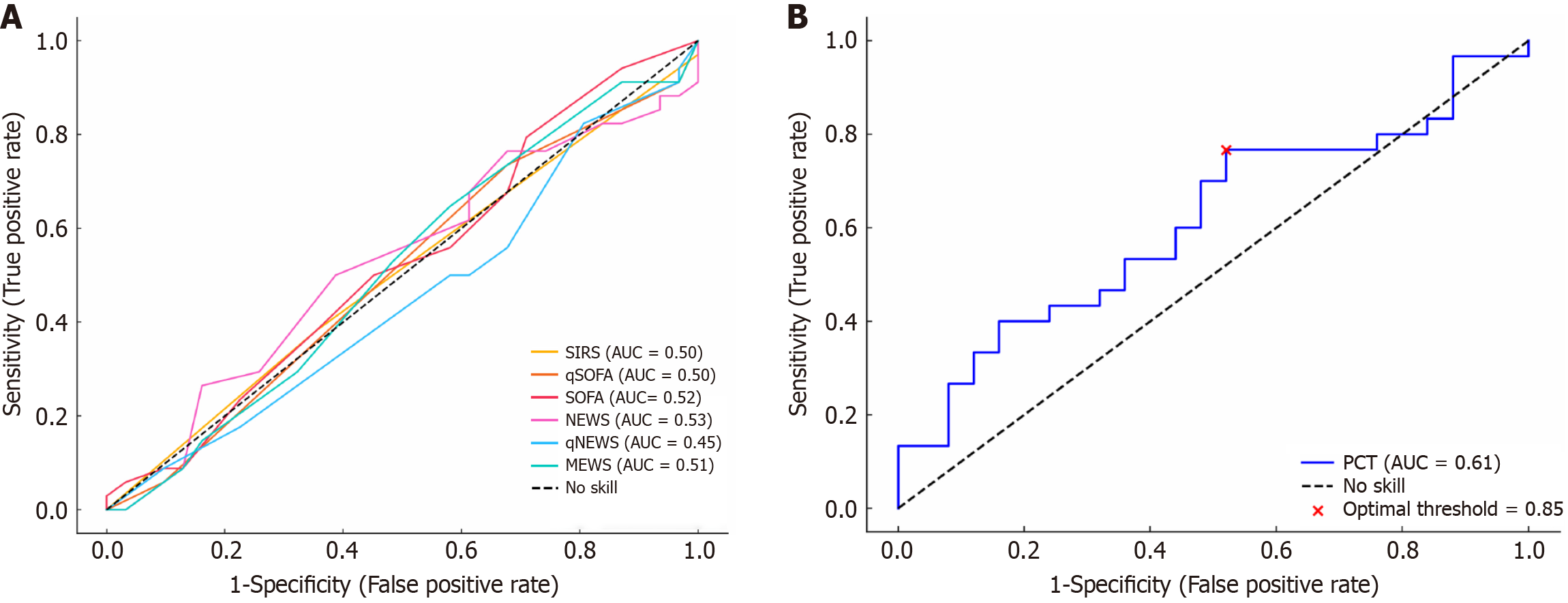
None of the sepsis screening tools at fever onset could reliably predict an infectious cause of nFUO, as their ROC curves were close to the diagonal reference line (AUC = 0.50), indicating poor discriminatory ability (Table 4, Figure 2A). Among sepsis biomarkers, serum PCT levels measured at fever onset were higher in the infectious group compared to the non-infectious group; however, the difference was not statistically significant (P = 0.156) (Table 4). Upon ROC analysis, PCT showed borderline predictive value [AUC = 0.61; 95% confidence interval (CI): 0.46-0.76] (Figure 2B). The optimal threshold was 0.85 μg/L, which performed slightly better than the conventional cutoff of 0.5 μg/L (Table 5). In contrast, CRP levels were paradoxically higher in the non-infectious cases, with an AUC of 0.45 (95%CI: 0.28-0.62), further limiting its diagnostic utility (Table 4). Combining PCT with sepsis tools such as SIRS or qSOFA did not improve predictive accuracy compared to PCT alone (Table 5).
| Parameter | Total (n = 80) | Infectious (n = 34) | Non-infectious (n = 31) | P value |
| Sepsis screening tools | ||||
| SIRS, median (IQR) | 3 (3-4) | 3 (3-4) | 3 (3-4) | 0.949 |
| SIRS ≥ 2, n (%) | 78 (97.5) | 33 (97.1) | 31 (100) | 1.000 |
| qSOFA, median (IQR) | 2 (1-2) | 2 (1-2) | 2 (1-2) | 0.957 |
| qSOFA ≥ 2, n (%) | 57 (71.3) | 25 (73.5) | 21 (67.7) | 0.608 |
| SOFA, median (IQR) | 6 (4-8) | 6.5 (4-8.3) | 6 (3-8) | 0.736 |
| NEWS, median (IQR) | 12 (9-14) | 12.5 (9.5-8.3) | 12 (8-14) | 0.726 |
| NEWS ≥ 7, n (%) | 67 (83.8) | 28 (82.4) | 27 (87.1) | 0.736 |
| qNEWS, median (IQR) | 6 (3-6) | 5 (3-6) | 6 (3-6) | 0.496 |
| MEWS, median (IQR) | 6 (4.25-8) | 7 (5-8) | 6 (5-8) | 0.863 |
| Sepsis biomarkers | ||||
| Procalcitonin (μg/L), median (IQR) | 1.9 (0.5-12.1) | 2.8 (0.8-25.9) | 1.0 (0.5-4.6) | 0.156 |
| C- reactive protein (mg/L), median (IQR) | 123.0 (55.9-225.6) | 107.2 (50.5-209.0) | 136.5 (51.9-245.3) | 0.566 |
| Parameters | PCT 0.85 μg/L | PCT 0.50 μg/L | qSOFA 2 + PCT 0.85 μg/L | SIRS 2 + PCT 0.85 μg/L |
| Sensitivity | 77% (58-90) | 77% (58-90) | 72% (51-88) | 76% (56-90) |
| Specificity | 48% (28-69) | 24% (9-45) | 44% (22-69) | 48% (28-69) |
| False negative rate | 23% (10-42) | 23% (10-42) | 28% (12-49) | 24% (10-44) |
| False positive rate | 52% (31-72) | 76% (55-91) | 56% (31-78) | 52% (31-72) |
| Positive likelihood ratio | 1.47 (0.96-2.26) | 1.01 (0.75-1.36) | 1.30 (0.80-2.09) | 1.46 (0.95-2.24) |
| Negative likelihood ratio | 0.49 (0.23-1.05) | 0.97 (0.38-2.52) | 0.63 (0.28-1.42) | 0.50 (0.23-1.08) |
| Diagnostic odds ratio | 3.03 (0.96-9.62) | 1.04 (0.30-3.69) | 2.06 (0.57-7.36) | 2.90 (0.91-9.23) |
| Youden’s index | 0.25 (0.00-0.49) | 0.01 (-0.22 to 0.23) | 0.16 (-0.12 to 0.45) | 0.24 (-0.01 to 0.49) |
Our nFUO cohort had an overall mortality rate of 20% (n = 16), with the highest mortality observed in the undiagnosed group (33.3%, n = 5) compared to the diagnosed cases (16.9%, n = 11); however, this difference was not statistically significant (P = 0.291). Among diagnosed cases, mortality rates were comparable between the infectious (n = 6, 17.6%) and non-infectious group (n = 5, 16.1%) (P = 0.185).
Fever resolution was achieved in 66 patients (82.5%), with no statistically significant difference between the infectious group (88.2%, n = 30), the non-infectious group (83.9%, n = 26), and the undiagnosed group (60%, n = 9) (P = 0.181). Fever duration was significantly longer in the infectious group (median: 7 days, IQR: 4.75-10.5) compared to the non-infectious (5 days, IQR: 4-6) and undiagnosed groups (4 days, IQR: 3-7) (P = 0.042). Prolonged fever (≥ 7 days) was observed in 55.9% of infectious cases (n = 19) vs 19.4% of non-infectious cases (n = 6) (P = 0.002). Hospital stay was also longest in the infectious group (median 19 days, IQR: 11.75-25) compared to the non-infectious (12 days, IQR: 8-19) and undiagnosed groups (12 days, IQR: 9-15) (P = 0.010).
This large prospective study evaluated the performance of sepsis screening tools and biomarkers in distinguishing infectious from non-infectious etiologies among critically ill patients with nFUO. Commonly used sepsis screening tools, including SIRS, qSOFA, SOFA, NEWS, qNEWS and MEWS, demonstrated poor diagnostic accuracy for infections, as indicated by ROC curves near the diagonal reference line. PCT showed modest-to-poor predictive value, with an AUC of 0.61 and an optimal threshold of 0.85 μg/L, but its overall performance remained limited. Combining PCT with sepsis screening tools did not improve diagnostic accuracy. In addition, CRP was an unreliable marker, paradoxically showing higher levels in non-infectious cases.
Understanding the etiology of nFUO is critical, as it varies across care settings (e.g., medical, surgical, oncology units) and significantly influences management strategies and clinical outcomes[2-5]. Our findings align with previous studies emphasizing the diagnostic complexity of nFUO, particularly in critically ill populations. Infectious causes, such as VAP/HAP and bloodstream infections, remained predominant[2-5]. However, their diagnosis often required extensive and repeated microbiological and radiological evaluations. Conversely, non-infectious causes - such as central fever, drug fever, pneumonitis, thrombophlebitis, or thrombosis - demanded heightened clinical suspicion, meticulous exclusion of infections, and careful interpretation of potential diagnostic clues[2-5,17,19,20]. The diagnostic challenge is further illustrated by the 18.8% of cases in our cohort that remained undiagnosed despite comprehensive evaluations.
Sepsis screening tools are widely used to identify severe infections and sepsis[9,21]. Tools like qSOFA, SIRS, and qNEWS allow rapid evaluations, while comprehensive scores such as SOFA, NEWS, and MEWS incorporate broader physiological parameters to predict prognosis[9,10,21,22]. However, in our study, these tools exhibited poor discriminatory performance for nFUO, as reflected by ROC curves close to the diagonal reference line. This highlights their limited applicability in complex conditions like nFUO, where inflammatory responses and physiological parameters (upon which these tools heavily rely) may be influenced by non-infectious factors such as comorbidities, intensive organ support (e.g., mechanical ventilation, vasopressors), drug reactions, or procedural complications[23,24]. Additionally, hospital-acquired infections often lack the classic signs of sepsis seen in community-acquired infections, further diminishing their effectiveness. The cross-sectional calculation of these scores at fever onset in our patients may also fail to capture the dynamic progression of sepsis or organ dysfunction that can evolve before or after an infection is identified[21]. While sepsis screening tools remain valuable for triaging patients in broader acute care settings, their role in guiding the diagnostic evaluation of nFUO is constrained.
PCT and CRP are integral components of the sepsis workup in hospitalized patients, aiding in differentiating infectious from non-infectious causes of fever and guiding antibiotic therapy decisions. While PCT is widely regarded as a reliable marker of bacterial infections, emerging evidence consistent with our study highlights its modest diagnostic utility[6,9,25-27]. Despite all infectious etiologies in our cohort being predominantly gram-negative bacterial infections, PCT showed modest-to-poor diagnostic performance (AUC = 0.61) with an optimal cutoff of 0.85 μg/L. In contrast, its widely used threshold of ≥ 0.5 μg/L had poor specificity. These findings emphasize the need for contextual interpretation of PCT values rather than absolute reliance.
The combination of PCT with sepsis screening tools such as qSOFA and SIRS did not enhance discriminatory performance in our study, further supporting evidence that these strategies add minimal value in nFUO[28,29]. Similarly, CRP demonstrated paradoxically higher levels in non-infectious cases, reinforcing its unreliability in distinguishing infectious causes in this setting. These findings highlight the limitations of PCT and CRP in nuanced contexts like nFUO, where a complex interplay of infectious and non-infectious factors may confound inflammatory markers. Our findings align with a growing body of evidence suggesting that PCT and CRP should not be used in isolation to diagnose bacterial infections in critically ill patients[9,25].
As demonstrated in our study, the accurate diagnosis of nFUO is vital due to its high mortality, with undiagnosed cases carrying even greater risks. This higher mortality in undiagnosed patients may reflect diagnostic uncertainty, leading to delayed or empiric treatments that failed to address the underlying pathology. It is also possible that some of these patients had atypical or rapidly progressive conditions that remained unrecognized despite standard evaluation, such as occult infections, non-resolving sepsis, life-threatening drug reactions, or immune-mediated disorders. Notably, many cases of nFUO could be prevented through timely diagnostic interventions and stringent infection control measures. Early evaluation of new-onset fever in critically ill patients typically prioritizes identifying infectious causes, as prompt antimicrobial treatment can substantially improve outcomes[6]. Our findings further highlight that infectious cases are often associated with delayed fever resolution and prolonged hospital stays, underscoring the necessity of a systematic and comprehensive diagnostic approach.
Traditional culture-based methods frequently fall short due to their time-consuming nature and limited sensitivity[30]. Emerging methods like metagenomic next-generation sequencing (mNGS) offer comprehensive pathogen detection and have shown the potential to reduce unnecessary antibiotic use in classical FUO[30,31]. Similarly, advanced imaging modalities like FDG-PET, well-established in classic FUO, are increasingly valuable in critically ill patients with unexplained fever or sepsis[2,6,32]. Novel sepsis biomarkers, such as presepsin, have also demonstrated superior diagnostic performance compared to PCT[33]. Given the limitations of existing tools such as sepsis scores, PCT, and CRP, future research should evaluate mNGS, FDG-PET imaging, and novel biomarkers to improve diagnostic accuracy for infectious causes of nFUO.
Our study has several limitations. Conducting the research in a single tertiary care center limits the generalizability of the findings to other healthcare settings, particularly those with different resources or patient demographics. The study population included critically ill patients without severe immunocompromised states or malignancies, further restricting the applicability of the results to general ward patients or oncology populations.
Although the sample size of 80 may appear modest, it was accrued over 18 months and represents one of the largest prospective datasets specifically focused on nFUO in critically ill adults. Given the diagnostic complexity and relatively low incidence of nFUO, this sample reflects the practical challenges of enrolling such a cohort. Nonetheless, we acknowledge that a larger sample size and inclusion of multiple centers would improve the external validity of the findings. Although infectious disorders were diagnosed based on standard Centers for Disease Control and Prevention and National Healthcare Safety Network definitions and nearly all cases (all but two) were microbiologically confirmed, the reliance on conventional culture-based diagnostic methods may have underestimated certain infectious etiologies, particularly fastidious or atypical pathogens[16,28]. Unassessed confounders, such as prior antimicrobial use - which can significantly affect microbiological culture yield - were not addressed[28].
While the diagnostic evaluation of new-onset fever adhered to recent SCCM/IDSA guidelines, FDG-PET could not be performed in some cases due to the unavailability of timely appointments or logistical challenges in shifting critically ill patients[6]. Additionally, the absence of a formal sample size calculation, while methodologically acceptable for exploratory diagnostic studies, limits the statistical power to detect modest associations. The use of convenience sampling based on a fixed study duration may also introduce selection bias and limit the external validity of our findings. Overall, the findings of this study should be considered exploratory and hypothesis-generating, warranting validation through larger, multicentric studies that include diverse populations and the integration of advanced diagnostic modalities.
Our study highlights the diagnostic challenges of nFUO in critically ill patients, demonstrating the limited utility of sepsis screening tools and biomarkers like PCT and CRP in reliably distinguishing infectious from non-infectious causes. Mortality remains high, particularly in undiagnosed cases, with infectious causes associated with delayed fever resolution and prolonged hospital stays. These findings emphasize the need for advanced diagnostic modalities, such as mNGS, FDG-PET, and novel biomarkers, to improve the early identification of infectious etiologies and enhance the diagnostic evaluation of nFUO.
| 1. | Durack DT, Street AC. Fever of unknown origin--reexamined and redefined. Curr Clin Top Infect Dis. 1991;11:35-51. [PubMed] |
| 2. | Haidar G, Singh N. Fever of Unknown Origin. N Engl J Med. 2022;386:463-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 93] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 3. | Abolnik IZ, Fahhoum JS, Cleveland KO, Abramson MA, Corey GR, Gelfand MS, Sexton DJ. Nosocomial Fever of Unknown Origin. Infect Dis Clin Pract. 1999;8:396-398. [RCA] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 4. | Al-Tonbary YA, Soliman OE, Sarhan MM, Hegazi MA, El-Ashry RA, El-Sharkawy AA, Salama OS, Yahya R. Nosocomial infections and fever of unknown origin in pediatric hematology/oncology unit: a retrospective annual study. World J Pediatr. 2011;7:60-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Engelhart S, Glasmacher A, Exner M, Kramer MH. Surveillance for nosocomial infections and fever of unknown origin among adult hematology-oncology patients. Infect Control Hosp Epidemiol. 2002;23:244-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | O'Grady NP, Alexander E, Alhazzani W, Alshamsi F, Cuellar-Rodriguez J, Jefferson BK, Kalil AC, Pastores SM, Patel R, van Duin D, Weber DJ, Deresinski S. Society of Critical Care Medicine and the Infectious Diseases Society of America Guidelines for Evaluating New Fever in Adult Patients in the ICU. Crit Care Med. 2023;51:1570-1586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 40] [Reference Citation Analysis (0)] |
| 7. | Dankul P, Karaketklang K, Jitmuang A. Nosocomial Fever in General Medical Wards: A Prospective Cohort Study of Clinical Characteristics and Outcomes. Infect Drug Resist. 2021;14:3873-3881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 8. | Pannu AK, Golla R, Kumari S, Suri V, Gupta P, Kumar R. Aetiology of pyrexia of unknown origin in north India. Trop Doct. 2021;51:34-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, Machado FR, Mcintyre L, Ostermann M, Prescott HC, Schorr C, Simpson S, Wiersinga WJ, Alshamsi F, Angus DC, Arabi Y, Azevedo L, Beale R, Beilman G, Belley-Cote E, Burry L, Cecconi M, Centofanti J, Coz Yataco A, De Waele J, Dellinger RP, Doi K, Du B, Estenssoro E, Ferrer R, Gomersall C, Hodgson C, Møller MH, Iwashyna T, Jacob S, Kleinpell R, Klompas M, Koh Y, Kumar A, Kwizera A, Lobo S, Masur H, McGloughlin S, Mehta S, Mehta Y, Mer M, Nunnally M, Oczkowski S, Osborn T, Papathanassoglou E, Perner A, Puskarich M, Roberts J, Schweickert W, Seckel M, Sevransky J, Sprung CL, Welte T, Zimmerman J, Levy M. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intensive Care Med. 2021;47:1181-1247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 295] [Cited by in RCA: 2860] [Article Influence: 572.0] [Reference Citation Analysis (0)] |
| 10. | Holland M, Kellett J. The United Kingdom's National Early Warning Score: should everyone use it? A narrative review. Intern Emerg Med. 2023;18:573-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 11. | Pannu AK, Saroch A, Kumar M, Behera A, Nayyar GS, Sharma N. Quantification of chronic diseases presenting in the Emergency Department and their disposition outcomes: A hospital-based cross-sectional study in north India. Trop Doct. 2022;52:276-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32099] [Cited by in RCA: 39738] [Article Influence: 1018.9] [Reference Citation Analysis (0)] |
| 13. | Charlson M, Szatrowski TP, Peterson J, Gold J. Validation of a combined comorbidity index. J Clin Epidemiol. 1994;47:1245-1251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4325] [Cited by in RCA: 5294] [Article Influence: 165.4] [Reference Citation Analysis (0)] |
| 14. | World Health Organization. International Classification of Diseases, Eleventh Revision (ICD-11) for Mortality and Morbidity Statistics. 2022. [cited 27 February 2025]. Available from: https://icdcdn.who.int/icd11referenceguide/en/refguide.pdf. |
| 15. | Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6072] [Cited by in RCA: 9461] [Article Influence: 630.7] [Reference Citation Analysis (1)] |
| 16. | United States Centers for Disease Control and Prevention. National Healthcare Safety Network (NHSN) Patient Safety Component Manual. January 2025. [cited 27 February 2025]. Available from: https://www.cdc.gov/nhsn/pdfs/pscmanual/pcsmanual_current.pdf. |
| 17. | Goyal K, Garg N, Bithal P. Central fever: a challenging clinical entity in neurocritical care. J Neurocrit Care. 2020;13:19-31. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Tenner S, Vege SS, Sheth SG, Sauer B, Yang A, Conwell DL, Yadlapati RH, Gardner TB. American College of Gastroenterology Guidelines: Management of Acute Pancreatitis. Am J Gastroenterol. 2024;119:419-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 208] [Article Influence: 104.0] [Reference Citation Analysis (0)] |
| 19. | Mandell LA, Niederman MS. Aspiration Pneumonia. N Engl J Med. 2019;380:651-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 408] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 20. | Naranjo CA, Busto U, Sellers EM, Sandor P, Ruiz I, Roberts EA, Janecek E, Domecq C, Greenblatt DJ. A method for estimating the probability of adverse drug reactions. Clin Pharmacol Ther. 1981;30:239-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7061] [Cited by in RCA: 8478] [Article Influence: 188.4] [Reference Citation Analysis (5)] |
| 21. | Seymour CW, Liu VX, Iwashyna TJ, Brunkhorst FM, Rea TD, Scherag A, Rubenfeld G, Kahn JM, Shankar-Hari M, Singer M, Deutschman CS, Escobar GJ, Angus DC. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315:762-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3076] [Cited by in RCA: 2667] [Article Influence: 266.7] [Reference Citation Analysis (0)] |
| 22. | McGrath SP, Perreard I, MacKenzie T, Calderwood M. Improvement of sepsis identification through multi-year comparison of sepsis and early warning scores. Am J Emerg Med. 2022;51:239-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Olander A, Frick L, Johansson J, Wibring K. The performance of screening tools and use of blood analyses in prehospital identification of sepsis patients and patients suitable for non-conveyance - an observational study. BMC Emerg Med. 2024;24:180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 24. | Kumar A, Pannu AK, Kumar M, Angrup A, Dutta P, Sharma N. Sepsis screening tools for predicting infection triggers and outcomes in diabetic ketoacidosis. Biomark Med. 2023;17:947-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 25. | Lawandi A, Oshiro M, Warner S, Diao G, Strich JR, Babiker A, Rhee C, Klompas M, Danner RL, Kadri SS. Reliability of Admission Procalcitonin Testing for Capturing Bacteremia Across the Sepsis Spectrum: Real-World Utilization and Performance Characteristics, 65 U.S. Hospitals, 2008-2017. Crit Care Med. 2023;51:1527-1537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Pannu AK, Kumar A, Kiran R, Bhatia M, Sharda SC, Saroch A, Angrup A, Dutta P, Sharma N. Diagnostic utility of procalcitonin for bacterial infections in diabetic ketoacidosis. Clin Exp Med. 2023;23:5299-5306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 27. | Demirdal T, Sen P, Nemli SA. Diagnostic Value of Procalcitonin in Predicting Bacteremia in Intensive Care Unit. Indian J Crit Care Med. 2018;22:78-84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Nandakumar A, Sudeep S, Sreemohan AC, Vijayakumar S, Sudhakaran GJ, Gutjahr G, Pathinaruporthi RK, Balachandran S, Chandra S, Purushothaman SS, Mohamed ZU, Nair SN, Moni M, Sathyapalan DT. Developing Augmented Pro-SOFA and Pro-SAPS Models by Integrating Biomarkers PCT, NLR, and CRP with SOFA and SAPS-III Scores. Indian J Crit Care Med. 2024;28:935-941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 29. | Yu H, Nie L, Liu A, Wu K, Hsein YC, Yen DW, Lee MG, Lee CC. Combining procalcitonin with the qSOFA and sepsis mortality prediction. Medicine (Baltimore). 2019;98:e15981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 30. | Neyton LPA, Langelier CR, Calfee CS. Metagenomic Sequencing in the ICU for Precision Diagnosis of Critical Infectious Illnesses. Crit Care. 2023;27:90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 31. | Marra AR, Lopes GOV, Pardo I, Hsieh MK, Kobayashi T, Marra PS, Marschall J, Pinho JRR, Amgarten DE, de Mello Malta F, Dos Santos NV, Edmond MB. Metagenomic next-generation sequencing in patients with fever of unknown origin: A comprehensive systematic literature review and meta-analysis. Diagn Microbiol Infect Dis. 2024;110:116465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 32. | van Leer B, van Rijsewijk ND, Nijsten MWN, Slart RHJA, Pillay J, Glaudemans AWJM. Practice of (18)F-FDG-PET/CT in ICU Patients: A Systematic Review. Semin Nucl Med. 2023;53:809-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 33. | He RR, Yue GL, Dong ML, Wang JQ, Cheng C. Sepsis Biomarkers: Advancements and Clinical Applications-A Narrative Review. Int J Mol Sci. 2024;25:9010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 67] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/