Published online Sep 9, 2022. doi: 10.5492/wjccm.v11.i5.317
Peer-review started: June 22, 2021
First decision: July 31, 2021
Revised: August 13, 2021
Accepted: July 5, 2022
Article in press: July 5, 2022
Published online: September 9, 2022
Processing time: 441 Days and 4.9 Hours
Intensive care unit (ICU) patients demand continuous monitoring of several clin
To develop a binary classifier for the outcome of death in ICU patients based on clinical and laboratory parameters, a set formed by 1087 instances and 50 vari
For categorical variables, frequencies and risk ratios were calculated. Numerical variables were computed as means and standard deviations and Mann-Whitney U tests were performed. We then divided the data into a training (80%) and test (20%) set. The training set was used to train a predictive model based on the Random Forest algorithm and the test set was used to evaluate the predictive effectiveness of the model.
A statistically significant association was identified between need for intubation, as well predominant systemic cardiovascular involvement, and hospital death. A number of the numerical variables analyzed (for instance Glasgow Coma Score punctuations, mean arterial pressure, temperature, pH, and lactate, creatinine, albumin and bilirubin values) were also significantly associated with death outcome. The proposed binary Random Forest classifier obtained on the test set (n = 218) had an accuracy of 80.28%, sensitivity of 81.82%, specificity of 79.43%, positive predictive value of 73.26%, negative predictive value of 84.85%, F1 score of 0.74, and area under the curve score of 0.85. The predictive variables of the greatest importance were the maximum and minimum lactate values, adding up to a predictive importance of 15.54%.
We demonstrated the efficacy of a Random Forest machine learning algorithm for handling clinical and laboratory data from patients under intensive monitoring. Therefore, we endorse the emerging notion that machine learning has great potential to provide us support to critically question existing methodologies, allowing improvements that reduce mortality.
Core Tip: Considering the critical nature of patients admitted to intensive care units (ICUs), this study seeks to analyze clinical and laboratory data using a machine learning model based on a Random Forest algorithm. Consequently, we developed a binary classifier that forecasts death outcome, achieving a relevant area under the curve value of 0.85 and identifying the variables that contributed the most to the prediction. With this, we aim to contribute to the improvement and methodological advancement in the development of clinically relevant machine learning tools, seeking to make medical practice decisions more accurate and reduce mortality in ICU patients.
- Citation: Caires Silveira E, Mattos Pretti S, Santos BA, Santos Corrêa CF, Madureira Silva L, Freire de Melo F. Prediction of hospital mortality in intensive care unit patients from clinical and laboratory data: A machine learning approach. World J Crit Care Med 2022; 11(5): 317-329
- URL: https://www.wjgnet.com/2220-3141/full/v11/i5/317.htm
- DOI: https://dx.doi.org/10.5492/wjccm.v11.i5.317
The intensive care unit (ICU) is the section of the hospital responsible for monitoring acute patients, and it relies on specialized multidisciplinary staff and high-technology equipment to ensure the best support for these patients, who are usually unstable and at high risk of death. These patients demand continuous monitoring of the most diverse clinical and laboratory parameters that directly influence their medical progress and the staff’s decision-making. Lactate levels obtained from arterial blood samples, for example, may indicate the levels and severity of tissue hypoxia[1]. The elevation in serum lactate levels (hyperlactatemia) is associated with increased mortality[2,3]. Another important parameter in critically ill patients is the prothrombin time expressed in international normalized ratio (INR), which reveals abnormalities in the coagulation status[4]. This parameter is also associated with an increased mortality when at altered levels. Besides these, many other laboratory and clinical data like temp
These data are so vital in the assistance of these patients that they are already used by several scoring systems, including the Acute Physiology and Chronic Health Evaluation (APACHE) and the Simplified Acute Physiology Score (SAPS), which are designed to assess and predict the patient’s prognosis and allow for appropriate interventions[6]. The APACHE score, for example, which has been widely used since its creation in the 1980s and has been undergoing updates ever since, relies on the use of para
Machine learning may be understood as a scientific discipline by which a computer system is enabled to cross-reference numerous data in order to build statistical prediction models through pattern re
The use of machine learning has been consolidated as an alternative for the development of predi
The limiting matter caused by the database used in machine learning predictive models was also observed in the study by Hou et al[12], who developed a model regarding 30-d mortality in patients who fit the Third International Consensus Definitions for Sepsis (Sepsis-3). This paper used a public database Medical Information Mart for Intensive Care III (MIMIC III) from a single-center critical care database. Another study that also relates the development of a predictive machine learning model in the context of patients with sepsis is the one proposed by Nemati et al[13] that, in addition to using the aforementioned MIMIC III, also relied on ICU admission data from two hospital centers. In this study, as well as in the two previously mentioned, the potentialuses of this tool in the early identification of severity of cases and the possibility of making fundamental decisions to the positive outcome for pat
In addition, more recently, in light of the advent of the severe acute respiratory syndrome coronavirus 2 pandemic, the application of these predictive models using machine learning technology have been employed on various grounds, such as for risk of critical coronavirus disease 2019 (COVID-19)[14], need for ICU transfer, and the prognosis of intensive care COVID-19 patients[15,16]. The latter one was associated with eight main component factors, namely: Lymphocyte percentage, prothrombin time, lactate dehydrogenase, total bilirubin, eosinophil percentage, creatinine, and neutrophil per
Understanding a clinical setting as complex and full of variables as the ICU, identifying existing patterns, and enabling outcome prediction is a valuable tool for the improvement of health assistance to these patients. Therefore, the aim of the current paper is to develop a predictive model for the outcome of death in ICU patients based on clinical and laboratory parameters using a binary classifier, with predicted outcome consisting of in-hospital death and discharge.
We used anonymized retrospective data from ICU patients admitted to the emergency department to build a predictive model geared towards predicting death outcomes in these patients. For this purpose, a dataset used in the study was created from the larger “WiDS (Women in Data Science) Datathon 2020: ICU Mortality Prediction” dataset[17], which presents clinical and laboratory data pertaining to the first 24 h of ICU patient admission. The criteria for inclusion of instances (i.e., patients) in the study dataset were: (1) ICU admission and emergency department admission; and (2) Completeness (i.e., absence of missing data) with respect to the variables of interest. Since all the data were obtained from a public and anonymized dataset[16], it was not necessary to submit this study to the ethics committee, being in accordance with all the established precepts by the Committee on Publication Ethics.
Aligned with the goal of building an interpretable predictive model from clinical and laboratory data, variables related to the clinical status of patients (such as vital signs, clinical score scores, blood counts, and biochemical test results) were prioritized in the definition of variables of interest - with exclusion of variables of this type only when redundant or when they represented the application of formulas instead of measured or scored values - to the detriment of anthropometric and demographic variables, with age being the only representative of this group of variables included. Additionally, factors referring to logistical aspects of hospitalization (such as source and type of admission and readmission status) were also not included among the variables of interest.
This way, a set formed by 1087 instances and 50 variables was obtained, of which 49 were assumed as predictive variables and 1 as predicted variable (outcome variable). The predictive numerical variables were: (1) Age; (2) Disease score; (3) Eye opening score on the Glasgow coma scale (GCS); (4) Heart rate; (5) Hematocrit; (6) Mean arterial pressure; (7) Maximum albumin; (8) Maximum bilirubin; (9) Maximum blood urea nitrogen; (10) Maximum calcium; (11) Maximum creatinine; (12) Maximum diastolic blood pressure; (13) Maximum glucose; (14) Maximum HCO3; (15) Maximum hemoglobin; (16) Maximum INR; (17) Maximum lactate; (18) Maximum platelets; (19) Maximum potassium; (20) Maximum sodium; (21) Minimum systolic blood pressure; (22) Maximum saturation of peripheral oxygen (SpO2); (23) Maximum white blood cells (WBC); (24) Minimum albumin; (25) Minimum bilirubin; (26) Maximum blood urea nitrogen; (27) Minimum calcium; (28) Minimum creatinine; (29) Minimum diastolic blood pressure; (30) Minimum glucose; (31) Minimum HCO3; (32) Minimum hemoglobin; (33) Minimum INR; (34) Minimum lactate; (35) Minimum platelets; (36) Minimum potassium; (37) Minimum sodium; (38) Minimum systolic blood pressure; (39) Minimum SpO2; (40) Minimum WBC; (41) Motor response on the GCS; (42) Partial PaO2; (43) Partial pressure of carbonic gas in arterial blood (PaCO2); (44) pH; (45) Respiratory rate; (46) Temperature; and (47) Verbal response on the GCS. The predictive categorical variables were: (1) Need for intubation or not; and (2) Predominant systemic involvement. The outcome variable was the evolution or not with hospital death.
The disease score corresponded to the number of diseases present among the following conditions: (1) Acquired immunodeficiency syndrome; (2) Cirrhosis; (3) Diabetes; (4) Hepatic failure; (5) Immunosuppression; (6) Leukemia; (7) Lymphoma; and (8) Solid tumor. The categories of predominant systemic involvement considered were: (1) Cardiovascular involvement; (2) Gastrointestinal involvement; (3) Genitourinary involvement; (4) Hematological involvement; (5) Metabolic involvement; (6) Musculoskeletal/skin involvement; (7) Neurological involvement; (8) Respiratory involvement; (9) Sepsis; and (10) Trauma.
Initially, a descriptive and comparative analysis of the data was performed. The data were categorized according to the outcome variable. After that, the occurrence frequencies of each category for of categorical predictive variables and the means and standard deviations for all numerical predictive variables in both groups were computed. Finally, the differences for each variable between the groups were analyzed using the χ2 test for risk ratios (for categorical variables) and the Mann-Whitney U test (for numerical variables). Since a decision tree ensemble algorithm was chosen to constitute our predictive model, it was not necessary to normalize or standardize the data, since tree partitioning algorithms are insensitive to scaling.
To perform our predictive analysis, we chose to build a Random Forest algorithm, a model consisting of an ensemble of randomized decision trees. As an extension of bootstrap aggregation (bagging) of decision trees, in Random Forest algorithms each individual model in the ensemble is employed to generate a prediction for a new sample, and these individual model predictions are averaged to give the forest’s prediction, resulting in better performance than any single tree. By combining individual models, the ensemble model tends to be more flexible and efficient. Accordingly, random forests have been incredibly successful in a variety of classification and regression problems with clinical applications. Furthermore, the algorithm does not require any feature scaling since decision trees predictions are partitioning-based instead of distance-based.
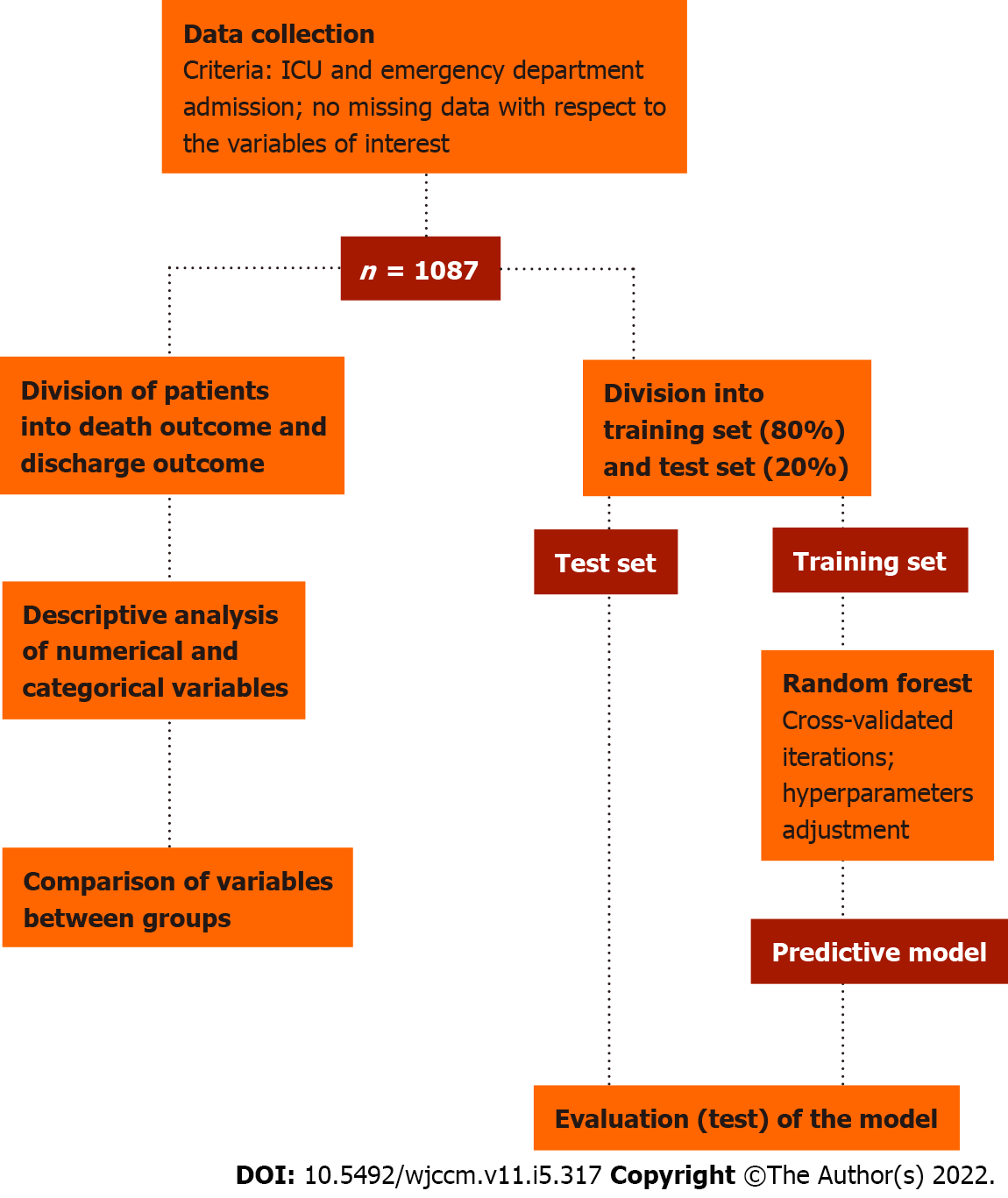
We then proceeded to the development of the predictive model for the outcome variable. The data were divided into a training set (80%) and a test set (20%). The training set was used to train a predictive model based on the Random Forest algorithm[18], implemented here through the Scikit-learn open source library[19]. The test set was used to evaluate the predictive effectiveness of the model. The metrics used for such evaluation were accuracy, sensitivity, specificity, area under the curve (AUC) score, positive predictive value, and negative predictive value. The adopted methodology is schematically summarized in Figure 1. Besides the predictive performance, the feature importance attributed by the model to each variable was also considered, which not only adds explainability to the model, but also potentially provides insights regarding the evaluation of critically ill patients and the factors associated with higher mortality in this clinical setting. All steps of statistical analysis and development of the predictive model were performed in Python (version 3.6.9) using SciPy and Scikit-learn libraries.
Data from 1087 ICU patients were analyzed and used in the construction of the predictive model, of which 388 evolved with hospital death, while the remaining 699 did not. With regard to the predictive variables categories - need or not of intubation and predominantly affected body system -, among the 388 patients who evolved with hospital death: 275 were intubated and 63 were not; 106 had sepsis as predominant systemic involvement, 18 respiratory involvement, 4 metabolic involvement, 154 cardiovascular involvement, 11 trauma, 16 neurological involvement, 25 gastrointestinal involvement, 2 genitourinary involvement, 1 musculoskeletal/skin involvement, and 1 hematological involvement. Among the 699 patients who did not progress to hospital death: 534 were intubated and 215 were not; 206 had sepsis as predominant systemic involvement, 107 respiratory involvement, 79 metabolic involvement, 167 cardiovascular involvement, 38 trauma, 49 neurological involvement, 74 gastro
| Variable | mean ± SD | U value | P value | |
| Death outcome, n = 338 | Survival outcome, n = 749 | |||
| Age | 63.4 ± 15.7 | 60.1 ± 16.1 | 111072 | < 0.001 |
| Disease score | 1.3 ± 0.8 | 1.2 ± 0.7 | 121505.5 | 0.110 |
| Eye opening (GCS) | 2.0 ± 1.2 | 2.5 ± 1.2 | 97325.0 | < 0.001 |
| Heart rate | 114.3 ± 34.9 | 111.1 ± 31.1 | 117672.0 | 0.031 |
| Hematocrit | 31.7 ± 8.3 | 32.8 ± 7.3 | 116749.0 | 0.02 |
| MAP | 84.7 ± 53.9 | 87.4 ± 48.7 | 108432.0 | < 0.001 |
| Max albumin | 2.7 ± 0.7 | 2.8 ± 0.6 | 109136.0 | < 0.001 |
| Max bilirubin | 2.2 ± 3.8 | 1.2 ± 1.8 | 98589.5 | < 0.001 |
| Max BUN | 40.0 ± 25.2 | 33.8 ± 24.5 | 102300.0 | < 0.001 |
| Max calcium | 8.0 ± 0.9 | 8.1 ± 0.8 | 117155.0 | 0.024 |
| Max creatinine | 2.6 ± 2.0 | 2.0 ± 1.9 | 96278.5 | < 0.001 |
| Max DBP | 92.0 ± 23.1 | 94.8 ± 21.5 | 116162.0 | 0.015 |
| Max glucose | 231.4 ± 113.0 | 210.1 ± 105.2 | 11090.0 | < 0.001 |
| Max HCO3 | 21.0 ± 5.1 | 23.3 ± 4.8 | 94750.0 | < 0.001 |
| Max hemoglobin | 11.7 ± 2.5 | 11.7 ± 2.3 | 124619.0 | 0.341 |
| Max INR | 2.1 ± 1.3 | 1.6 ± 0.8 | 83944.0 | < 0.001 |
| Max lactate | 7.3 ± 5.5 | 3.2 ± 2.8 | 62255.5 | < 0.001 |
| Max platelets | 189446.7 ± 98687.9 | 198186.9 ± 96842.7 | 120773.5 | 0.113 |
| Max potassium | 4.7 ± 0.9 | 4.5 ± 0.8 | 106603.0 | < 0.001 |
| Max sodium | 142.1 ± 6.7 | 140.9 ± 5.4 | 113894.0 | 0.004 |
| Max SBP | 147.5 ± 29.3 | 151.1 ± 26.2 | 113747.5 | 0.004 |
| Max SpO2 | 99.6 ± 1.5 | 99.8 ± 0.6 | 119714.5 | 0.005 |
| Max WBC | 17442.9 ± 10269.3 | 15302 ± 8516 | 111218.5 | 0.001 |
| Min albumin | 2.5 ± 0.7 | 2.7 ± 0.6 | 101997.5 | < 0.001 |
| Min bilirubin | 1.9 ± 3.3 | 1.1 ± 1.7 | 101177.5 | < 0.001 |
| Min BUN | 34.2 ± 22.9 | 29.0 ± 21.0 | 106584.5 | < 0.001 |
| Min calcium | 7.4 ± 0.9 | 7.7 ± 0.9 | 98668.5 | < 0.001 |
| Min creatinine | 2.08 ± 1.7 | 1.6 ± 1.3 | 99935.0 | < 0.001 |
| Min glucose | 104.6 ± 47.0 | 110.7 ± 38.1 | 111941.5 | 0.001 |
| Min HCO3 | 17.0 ± 5.5 | 20.6 ± 5.5 | 79747.5 | < 0.001 |
| Min hemoglobin | 10.3 ± 2.7 | 10.8 ± 2.4 | 111370.0 | 0.001 |
| Min INR | 1.8 ± 0.9 | 1.5 ± 0.6 | 89909.5 | < 0.001 |
| Min lactate | 4.7 ± 4.0 | 2.1 ± 1.58 | 69894.5 | < 0.001 |
| Min platelets | 157252 ± 94655.6 | 177120.8 ± 90595.7 | 110075.0 | < 0.001 |
| Min potassium | 3.8 ± 0.8 | 3.8 ± 0.7 | 125962.0 | < 0.001 |
| Min SBP | 75.4 ± 20.3 | 84.9 ± 19.6 | 92919.0 | < 0.001 |
| Min sodium | 137.9 ± 6.1 | 138.2 ± 5.5 | 121722.5 | 0.155 |
| Min WBC | 13247.1 ± 8505.4 | 12.7 ± 6.9 | 122208.5 | 0.181 |
| Min DBP | 38.7 ± 14.9 | 44.9 ± 12.7 | 93559.5 | < 0.001 |
| Min SpO2 | 81.3 ± 19.0 | 88.0 ± 12.0 | 94624.5 | < 0.001 |
| Motor response (GCS) | 2.9 ± 2.2 | 4.3 ± 2.0 | 83488.5 | < 0.001 |
| PaCO2 | 40.0 ± 13.9 | 39.5 ± 11.6 | 124352.0 | 0.321 |
| PaO2 | 137.4 ± 102.3 | 130.9 ± 82.4 | 121043.0 | 0.124 |
| pH | 7.3 ± 0.1 | 7.3 ± 0.1 | 109784.0 | < 0.001 |
| Respiratory rate | 31.2 ± 15.1 | 27.5 ± 14.9 | 107284.5 | < 0.001 |
| Temperature | 35.2 ± 1.9 | 36.2 ± 1.4 | 80674.5 | < 0.001 |
| Verbal response (GCS) | 1.9 ± 1.5 | 2.3 ± 1.7 | 109666.5 | < 0.001 |
The search for the best hyperparameters in our Random Forest model training was done using randomized search. In this way, 100 random combinations of hyperparameters were tested. Each combination was iterated 6 times, as a 6-fold validation scheme was adopted. In this scheme, the training set (n = 869) was split into 6 parts, and in each iteration a different part was used for validation. Ultimately, during training we performed 600 fits, obtaining the following hyperparameters: (1) Number of estimators = 213; (2) Maximum depth = 23; (3) Maximum leaf nodes = 24; (4) Minimum samples split = 5; (5) Class weights = 3.9; and (6) Bootstrap = true.
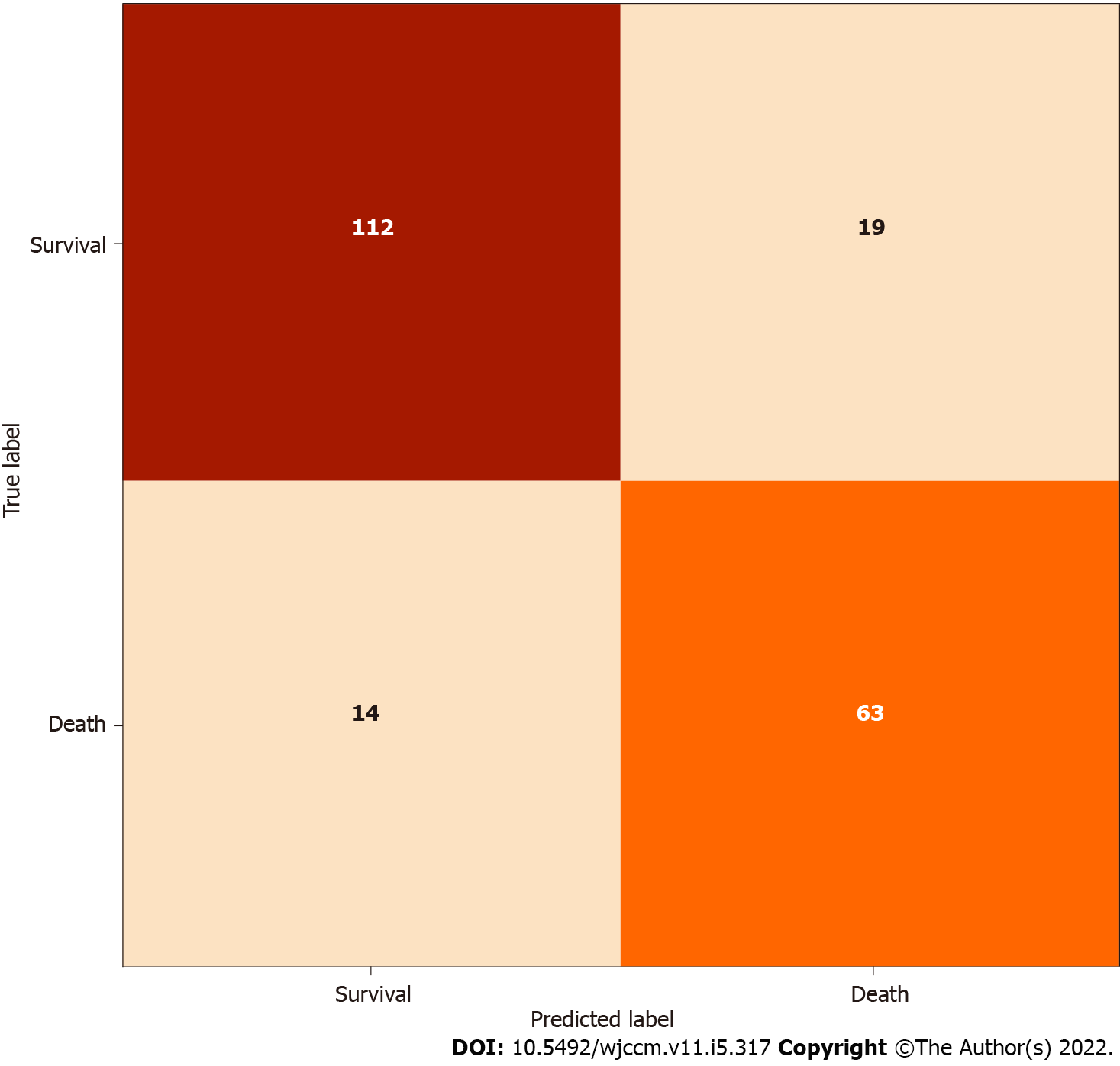
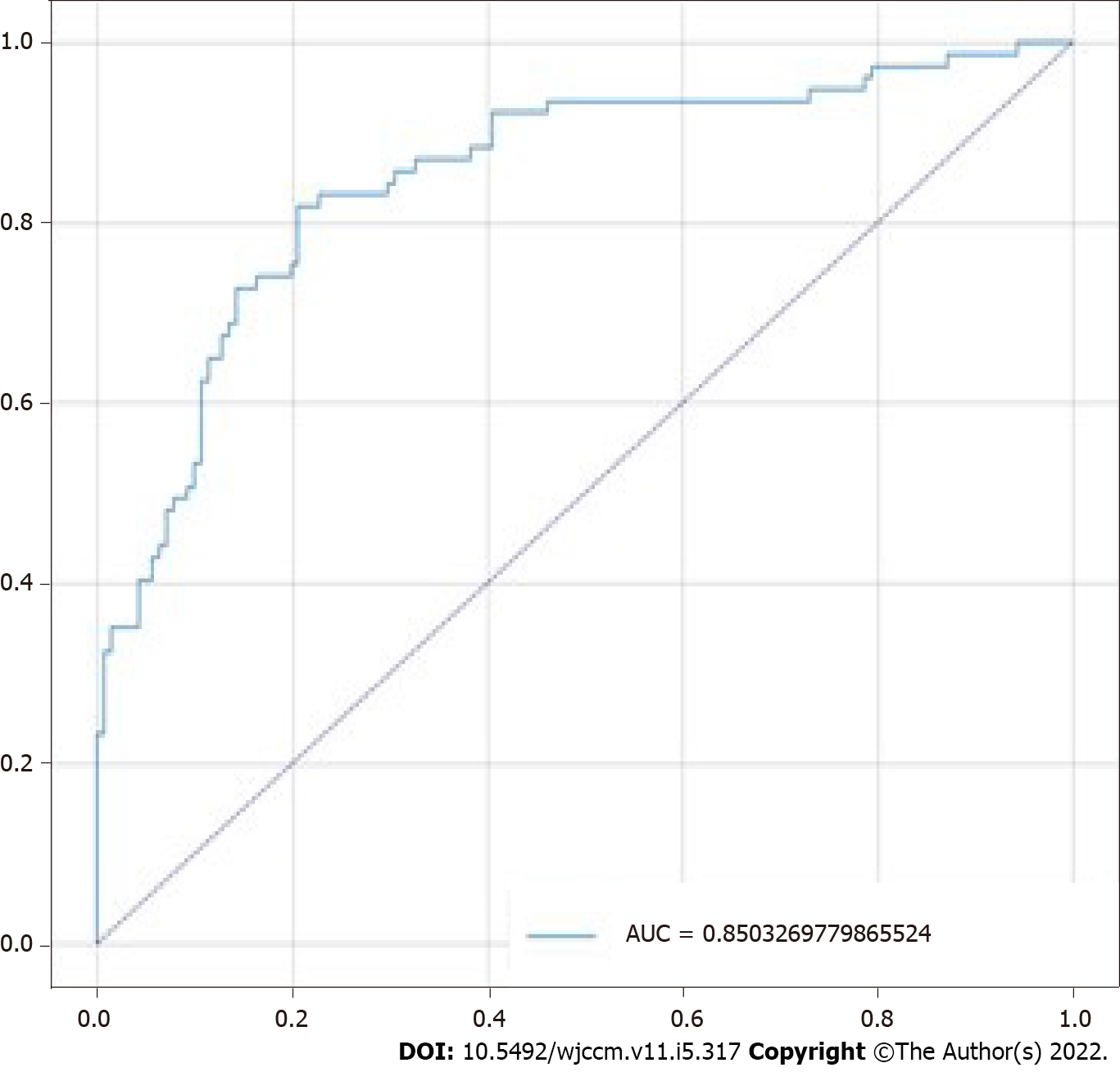
The model obtained accuracy of 80.28%, sensitivity of 81.82%, specificity of 79.43%, positive predictive value of 73.26%, negative predictive value of 84.85%, F1 score of 0.74, and AUC score of 0.85 on the test set (n = 218). The confusion matrix for the model is shown in Figure 2, and its receiver operating characteristic (ROC) curve is shown in Figure 3. The predictive variables with the greatest importance were the maximum and minimum lactate values, adding up to a predictive importance of 15.54%, followed by temperature (6.47%), motor punctuation in GCS (5.25%), maximum blood urea nitrogen (4.35%), and minimum WBC (3.31%). The percentage importance of the other variables in the prediction are listed in Table 2.
| Variable | Predictive importance, % |
| Maximum lactate | 9.05 |
| Minimum lactate | 6.49 |
| Temperature | 6.47 |
| Motor GCS | 5.25 |
| Maximum BUN | 4.35 |
| Minimum WBC | 3.31 |
| Minimum creatinine | 3.22 |
| Maximum INR | 3.15 |
| Minimum HCO3 | 2.84 |
| Maximum glucose | 2.69 |
| Minimum SpO2 | 2.45 |
| pH | 2.18 |
| Age | 2.09 |
| Minimum INR | 1.95 |
| Platelets | 1.9 |
| Maximum HCO3 | 1.83 |
| Minimum SBP | 1.82 |
| Minimum DBP | 1.82 |
| Maximum creatinine | 1.79 |
| Minimum albumin | 1.67 |
| Minimum sodium | 1.66 |
| Predominant systemic involvement | 1.64 |
| Maximum bilirubin | 1.63 |
| Maximum WBC | 1.63 |
| PaO2 | 1.62 |
| Minimum hemoglobin | 1.6 |
| Maximum SBP | 1.6 |
| Maximum albumin | 1.5 |
| MAP | 1.5 |
| Eyes opening GCS | 1.46 |
| Respiratory rate | 1.41 |
| Minimum calcium | 1.4 |
| Maximum hemoglobin | 1.39 |
| Minimum platelets | 1.35 |
| Minimum BUN | 1.28 |
| Hematocrit | 1.22 |
| Minimum bilirubin | 1.2 |
| PaCO2 | 1.19 |
| Maximum sodium | 1.13 |
| Maximum DBP | 1.12 |
| Maximum calcium | 0.93 |
| Minimum glucose | 0.92 |
| Minimum potassium | 0.92 |
| Maximum potassium | 0.82 |
| Heart rate | 0.72 |
| Verbal GCS | 0.42 |
| Intubated | 0.15 |
| Disease score | 0.14 |
| Maximum SpO2 | 0.13 |
The presented predictive model, a Random Forest binary classifier, was able to predict in the test set the occurrence or not of hospital death with an accuracy of 80.28%, sensitivity of 81.82%, and specificity of 79.43%. It is well established in the literature that this type of classifier is generally well suited for high-dimensional problems with highly correlated features (a frequent situation when it comes to medical data)[20]. Our results are consistent with that, as they demonstrate the potential for using random forests to handle clinical and laboratory data from patients under intensive monitoring.
The ICU mortality is high, and the patients require interventions that are cost-effective in order to avoid mortality without inputting unnecessary costs or demand to the medical team. Mortality prediction models work with the objective to assess the severity of the patients so that, based on its findings, the treatment needed can be directed. The analysis presented in this study works in the same way; if we identify those patients that have major mortality rates, faster and better care can be provided in order to prevent the worse outcome[21]. For this purpose, a variety of assessment scores already exist, like APACHE, SAPS or Mortality Probability Model (MPM). The ROC value of our model (0.85) was comparable with some of these highly used models, like 0.836 for APACHE II, or 0.826 for SAPS II[22], which showcase the good results obtained.
Furthermore, the machine learning approach to predict mortality in ICU patients has been documented. For example, Veith and Steele[23] developed a LazyKStar model to predict mortality in ICU patients at the time of hospital admission, obtaining a 10-fold validation AUC value of 0.75.A recurrent neural network inputted with 44 clinical and laboratory features from the first 24 h of ICU patient admission proposed by Thorsen-Meyer et al[24] achieved an AUC of 0.82. The extreme gradient boosted trees classifier developed by Chia et al[25] reached an AUC of 0.83 using 42 predictive variables. The formats and results of these last two studies are comparable to ours, since we reached an AUC of 0.85 using a random forest fed by 50 features.
Due to the COVID-19 pandemic, there was a great growth of publications focused on machine learning models for predicting ICU mortality in a disease-specific manner, such as those by Pan et al[16], Lichtner et al[26], and Subudhi et al[27]. Meanwhile, many of the previous studies in this field also focus on predicting ICU outcomes for specific diseases or morbid conditions, like sepsis or death from pulmonary tuberculosis[11,13,28], which lead to an assessment of parameters specific for the disease studied, somewhat restricting the research. Many of the renowned models and scales for ICU mortality prediction demand a series of measurements to make their use possible, but not always all the data required are available. In this sense, it is important to understand what the main variables involved related to the outcome of interest (and its prediction) are, so that they can be closely monitored. In our study, lactate level proved to be the most influential one, which is in accordance with its physiological role that indicates poor oxygenation, anaerobic metabolism, acidosis and muscle fatigue, involved in a systemic response of the organ is mand corroborates the findings by Bou Chebl et al[29], Villar et al[30] and Vincent et al[2]. Despite its predictive importance found in our study (15.54%), lactate is not a variable of most scores used, and is not included in APACHE, SAPS or MPM.
Temperature, which is part of APACHE and SAPS, was the second variable that influenced the most the outcome prediction; its variation (hyper or hypothermia) is related with a loss of control of body homeostasis, and the mean valor for death outcome was 35.2 ± 1.9. While we have an increase of nearly 1 point in the mean value for the survival outcome, these data could represent that an increase of the temperature or even fever could be a positive body response, indicating an immune system attempt to fight the pathology[31,32].
The third variable of major impact is the motor GCS punctuation, which is part of GCS, a widely known scale for neurologic damage used in hospital admissions as well as assessment models[33].This motor element has a specific field only in APACHE IV. Lower punctuations in GCS are related with greater neurologic damage, with 3 and 1 as its bottom punctuation for the global and motor scale respectively, the mean of 2.9 ± 2.2 for the death outcome in contrast with the value of 4.3 ± 2.0 for the survival mean demonstrate a considerable difference between those patients since the greatest value possible for the motor component is 6. The stratification of the data based on its predictive value is a great contribution since the variables above discussed account for approximately 27% of the result, while the other 45 for the remaining 73%, indicating that continuous monitoring of them may be of great value. Considering their importance, a detailed survey with either a dataset with per hour measurement of parameters or the data separated by ICU type could lead to more specific approaches for the medical staff.
Despite the good results found, this study faces as its main limitation the incompleteness of the original dataset for many instances regarding important clinical and laboratory variables, which lead to the use of a relatively small quantity of instances to train the predictive model. Since machine learning algorithms are essentially data-driven, a larger amount of data could lead to greater accuracy and a wider generalizability of the model, thus being useful for additional testing and refinement. Another potential limitation is related to the clinically broad nature of the variables analyzed, since the purpose was to study the possible parameters available in the ICU, which contrasts with research focused on the outcomes for a specific disease and, therefore, fed with more specific variables with regards to the considered pathophysiological process.
Although the use of a wide range of clinical and laboratory parameters was critical for our purpose of assessing the predictive significance of the variables in the context of building a model that is not only explainable but also clinically interpretable, this factor may restrict the possibilities of potential datasets to be used to ascertain the reproducibility of the findings, since some parameters may be unavailable. However, since these are variables commonly evaluated in critically ill patients in the ICU, for whom the prognostic evaluation of mortality is more important (in view of their higher mortality rates), we believe that this should not be a limiting factor to the clinical applicability of the proposed model.
In the study, it was possible to develop a reliable model for predicting mortality in the ICU, in which the influence of lactate level stands out as the main variable involved in the outcome prediction, followed by temperature and motor GCS. What can be perceived through the research is that machine learning comes to contribute and to make medical practice more efficient, as it allows faster analysis that otherwise would be complex and time-consuming. More than that, it also allows us to critically question existing parameters and methodologies through the results it provides in order to allow improvements that reduce the mortality of patients and are time and cost-effective. This study also highlights the importance of complete and organized registers of ICU patient data in order to enable the development of predictive models towards prevention and prediction of in-hospital bad outcomes.
The monitoring of clinical and laboratory parameters of patients in the intensive care unit (ICU) is an extremely important part of the routine of intensive care staff. Additionally, several scores already utilize these parameters to guide the assistance of these patients. In the meantime, the advance of technological resources, such as the machine learning approach, allows the development of predictive models capable of being applied to medical practice.
Mortality in the ICU is something that worries and drives the search for alternatives that can help the team in directing treatment to avoid this negative outcome. Therefore, a predictive model that uses the patient’s parameters can precisely influence this treatment guidance, improving the cost-effectiveness quickly and safely.
The objective of our study is the development of a binary classifier predictive model between the outcomes of death and non-death in ICU patients. This paper demonstrates the potency of emerging technological realities within the medical field and how it is possible to harness them to improve healthcare practices.
Initially, we obtained a set of 1087 instances and 50 variables related to patients admitted to an ICU by using a public database. We calculated frequency and risk rate for categorical variables and means, standard deviations, and the Mann-Whitney U test for numerical variables. Afterwards, we divided the data for the application in training of the predictive model based on the Random Forest algorithm and then to test the effectiveness of the model.
Among the 50 variables associated with death outcome, the maximum and minimum lactate values were the most important predictors (15.54%) followed by temperature (6.47%), and motor Glasgow coma scale punctuation (5.25%). The Random Forest binary classifier predictive model (death and no death) showed accuracy of 80.28%, sensitivity of 81.82%, specificity of 79.43%, positive predictive value of 73.26%, negative predictive value of 84.85%, F1 score of 0.74, and area under the curve score of 0.85.
This study demonstrated the development of a predictive model with high accuracy, sensitivity, and specificity for ICU patients by applying a machine learning approach, the Random Forest algorithm, to clinical and laboratory data.
The proper registration of patient parameters, as well as the availability of more and larger databases and even further development of digital tools, can enhance machine learning approaches, enabling the refinement of predictive models and patient care.
| 1. | Fuller BM, Dellinger RP. Lactate as a hemodynamic marker in the critically ill. Curr Opin Crit Care. 2012;18:267-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 119] [Article Influence: 10.8] [Reference Citation Analysis (1)] |
| 2. | Vincent JL, Quintairos E Silva A, Couto L Jr, Taccone FS. The value of blood lactate kinetics in critically ill patients: a systematic review. Crit Care. 2016;20:257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 378] [Cited by in RCA: 374] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 3. | Nichol AD, Egi M, Pettila V, Bellomo R, French C, Hart G, Davies A, Stachowski E, Reade MC, Bailey M, Cooper DJ. Relative hyperlactatemia and hospital mortality in critically ill patients: a retrospective multi-centre study. Crit Care. 2010;14:R25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 261] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 4. | Levi M, Opal SM. Coagulation abnormalities in critically ill patients. Crit Care. 2006;10:222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 209] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 5. | Hunt BJ. Bleeding and coagulopathies in critical care. N Engl J Med. 2014;370:847-859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 189] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 6. | Ko M, Shim M, Lee SM, Kim Y, Yoon S. Performance of APACHE IV in Medical Intensive Care Unit Patients: Comparisons with APACHE II, SAPS 3, and MPM0 III. Acute Crit Care. 2018;33:216-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 7. | Ihnsook J, Myunghee K, Jungsoon K. Predictive accuracy of severity scoring system: a prospective cohort study using APACHE III in a Korean intensive care unit. Int J Nurs Stud. 2003;40:219-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 8. | Deo RC. Machine Learning in Medicine. Circulation. 2015;132:1920-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1155] [Cited by in RCA: 2162] [Article Influence: 216.2] [Reference Citation Analysis (7)] |
| 9. | Heo J, Yoon JG, Park H, Kim YD, Nam HS, Heo JH. Machine Learning-Based Model for Prediction of Outcomes in Acute Stroke. Stroke. 2019;50:1263-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 380] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 10. | Lynch CM, Abdollahi B, Fuqua JD, de Carlo AR, Bartholomai JA, Balgemann RN, van Berkel VH, Frieboes HB. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int J Med Inform. 2017;108:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 140] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 11. | Liu Q, Gao J, Luo B, Liu J, Zhang L, Kang W, Han F. Prediction model for death in patients with pulmonary tuberculosis accompanied by respiratory failure in ICU: retrospective study. Ann Palliat Med. 2020;9:2731-2740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | Hou N, Li M, He L, Xie B, Wang L, Zhang R, Yu Y, Sun X, Pan Z, Wang K. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: a machine learning approach using XGboost. J Transl Med. 2020;18:462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 349] [Article Influence: 58.2] [Reference Citation Analysis (1)] |
| 13. | Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, Buchman TG. An Interpretable Machine Learning Model for Accurate Prediction of Sepsis in the ICU. Crit Care Med. 2018;46:547-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 450] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 14. | Assaf D, Gutman Y, Neuman Y, Segal G, Amit S, Gefen-Halevi S, Shilo N, Epstein A, Mor-Cohen R, Biber A, Rahav G, Levy I, Tirosh A. Utilization of machine-learning models to accurately predict the risk for critical COVID-19. Intern Emerg Med. 2020;15:1435-1443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 121] [Article Influence: 20.2] [Reference Citation Analysis (1)] |
| 15. | Cheng FY, Joshi H, Tandon P, Freeman R, Reich DL, Mazumdar M, Kohli-Seth R, Levin M, Timsina P, Kia A. Using Machine Learning to Predict ICU Transfer in Hospitalized COVID-19 Patients. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (1)] |
| 16. | Pan P, Li Y, Xiao Y, Han B, Su L, Su M, Zhang S, Jiang D, Chen X, Zhou F, Ma L, Bao P, Xie L. Prognostic Assessment of COVID-19 in the Intensive Care Unit by Machine Learning Methods: Model Development and Validation. J Med Internet Res. 2020;22:e23128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 17. | Lee M, Raffa J, Ghassemi M, Pollard T, Kalanidhi S, Badawi O, Matthys K, Celi LA. WiDS (Women in Data Science) Datathon 2020: ICU Mortality Prediction (version 1.0.0). PhysioNet. 2020;. [DOI] [Full Text] |
| 18. | Breiman L. Random Forests. Mach Learn. 2001;45:5-32. [RCA] [DOI] [Full Text] [Cited by in Crossref: 56052] [Cited by in RCA: 36453] [Article Influence: 2804.1] [Reference Citation Analysis (0)] |
| 19. | Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Louppe G, Prettenhofer P, Weiss R, Weiss RJ, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine Learning in Python. J MachLearn Res. 2011;12:2825-2830. |
| 20. | Yang F, Wang HZ, Mi H, Lin CD, Cai WW. Using random forest for reliable classification and cost-sensitive learning for medical diagnosis. BMC Bioinformatics. 2009;10 Suppl 1:S22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Awad A, Bader-El-Den M, McNicholas J, Briggs J, El-Sonbaty Y. Predicting hospital mortality for intensive care unit patients: Time-series analysis. Health Informatics J. 2020;26:1043-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Fuchs PA, Czech IJ, Krzych ŁJ. The Pros and Cons of the Prediction Game: The Never-ending Debate of Mortality in the Intensive Care Unit. Int J Environ Res Public Health. 2019;16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Veith N, Steele R. Machine Learning-based Prediction of ICU Patient Mortality at Time of Admission. Proceedings of the 2nd International Conference on Information System and Data Mining; 2018 Mar; Lakeland, USA. New York: Association for Computing Machinery, 2018: 34-38. [DOI] [Full Text] |
| 24. | Thorsen-Meyer HC, Nielsen AB, Nielsen AP, Kaas-Hansen BS, Toft P, Schierbeck J, Strøm T, Chmura PJ, Heimann M, Dybdahl L, Spangsege L, Hulsen P, Belling K, Brunak S, Perner A. Dynamic and explainable machine learning prediction of mortality in patients in the intensive care unit: a retrospective study of high-frequency data in electronic patient records. Lancet Digit Health. 2020;2:e179-e191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 184] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 25. | Chia A, Khoo M, Lim A, Ong K, Sun Y, Nguyen B, Chua M, Pang J. Explainable machine learning prediction of ICU mortality. Inform Med Unlocked. 2021;25:100674. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Lichtner G, Balzer F, Haufe S, Giesa N, Schiefenhövel F, Schmieding M, Jurth C, Kopp W, Akalin A, Schaller SJ, Weber-Carstens S, Spies C, von Dincklage F. Predicting lethal courses in critically ill COVID-19 patients using a machine learning model trained on patients with non-COVID-19 viral pneumonia. Sci Rep. 2021;11:13205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 27. | Subudhi S, Verma A, Patel AB, Hardin CC, Khandekar MJ, Lee H, McEvoy D, Stylianopoulos T, Munn LL, Dutta S, Jain RK. Comparing machine learning algorithms for predicting ICU admission and mortality in COVID-19. NPJ Digit Med. 2021;4:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 28. | Su L, Xu Z, Chang F, Ma Y, Liu S, Jiang H, Wang H, Li D, Chen H, Zhou X, Hong N, Zhu W, Long Y. Early Prediction of Mortality, Severity, and Length of Stay in the Intensive Care Unit of Sepsis Patients Based on Sepsis 3.0 by Machine Learning Models. Front Med (Lausanne). 2021;8:664966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 29. | Bou Chebl R, El Khuri C, Shami A, Rajha E, Faris N, Bachir R, Abou Dagher G. Serum lactate is an independent predictor of hospital mortality in critically ill patients in the emergency department: a retrospective study. Scand J Trauma Resusc Emerg Med. 2017;25:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 30. | Villar J, Short JH, Lighthall G. Lactate Predicts Both Short- and Long-Term Mortality in Patients With and Without Sepsis. Infect Dis (Auckl). 2019;12:1178633719862776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 31. | Young PJ, Saxena M, Beasley R, Bellomo R, Bailey M, Pilcher D, Finfer S, Harrison D, Myburgh J, Rowan K. Early peak temperature and mortality in critically ill patients with or without infection. Intensive Care Med. 2012;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 32. | Rehman T, deBoisblanc BP. Persistent fever in the ICU. Chest. 2014;145:158-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed
Peer-review model: Single blind
Specialty type: Critical care medicine
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cabezuelo AS, Spain S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Wang JJ