Peer-review started: May 20, 2015
First decision: October 17, 2015
Revised: January 19, 2015
Accepted: February 14, 2016
Article in press: February 16, 2016
Published online: March 27, 2016
Processing time: 314 Days and 17.6 Hours
AIM: To study the bacteriocidal or bacteriostatic role of mast cells during infection with Mycobacterium.
METHODS: Mycobacterium marinum (M. marinum) (BAA-535/M strain) was investigated for its ability to grow at a temperature relevant to the mammalian host. Primary mast cells were differentiated from bone marrows of mice, a human mast cell line (HMC-1) and a human monocytic cell line (MonoMac6) were maintained in culture. Mice were stimulated by intraperitoneal injection of heat-killed M. marinum to study cytochemically the degranulation of peritoneal mast cells. HMC-1 cells were stimulated with M. marinum to analyse mRNA expression for inflammatory reactant genes, while HMC-1 and primary mouse mast cells were infected with M. marinum to establish in parallel cell viability (lactate dehydrogenase release and cell counts) and viable mycobacterial counts. Flow cytometry was used to assess intracellular presence of fluorescein isothiocyanate labelled M. marinum after trypan blue quenching and to measure the extent of infection-induced apoptosis or necrosis in HMC-1. A GFP expressing recombinant M. marinum strain was used to assess intracellular location by fluorescence microscopy. Light microscopy of osmium tetroxide and Gram Twort stained sections of 0.5 μm and transmission electron microscopy were undertaken as sensitive methods.
RESULTS: Since its isolation, M. marinum has adapted to grow at 37 °C. This study found that M. marinum infects HMC-1 cells and primary murine mast cells, where they survive, replicate, and cause dose dependent cell damage over the analysis period of up to 120 h. Amikacin was an effective aminoglycoside antibiotic to eliminate extracellular or membrane attached M. marinum in order to adequately quantify the intracellular bacterial loads. In vivo, intraperitoneal injection of heat-killed M. marinum led to the release of mast cell granules in mice. HMC-1 cells stimulated with M. marinum showed a biphasic pattern of increased mRNA expression for LL-37 and COX-2/TNF-α during 24 h of stimulation. In HMC-1, M. marinum localised to the cytoplasm whereas in primary mast cells, M. marinum were found in vacuoles.
CONCLUSION: The effector role of mast cells in infection with M. marinum can be studied in vitro and in vivo.
Core tip: Mycobacterium marinum (M. marinum) is easily culturable and shows promise as a model to understand in mammalian cells the pathogenicity of M. tuberculosis. We used M. marinum to study uptake and elimination of M. marinum by mast cells, being abundant immune effector cells. A range of imaging techniques was used to unequivocally show the intracellular presence of M. marinum. Mast cells did not control the replication of M. marinum but reacted in a pro-inflammatory way. This is consistent with mast cells being orchestrators of inflammation. In summary, we clearly show that M. marinum can infect mast cells, survive and replicate within.
- Citation: Siad S, Byrne S, Mukamolova G, Stover C. Intracellular localisation of Mycobacterium marinum in mast cells. World J Immunol 2016; 6(1): 83-95
- URL: https://www.wjgnet.com/2219-2824/full/v6/i1/83.htm
- DOI: https://dx.doi.org/10.5411/wji.v6.i1.83
Mycobacterium marinum (M. marinum) is the causative agent of mycobacterial disease in poikilotherms and of the so-called “swimming pool” granuloma contracted by the human host[1]. Because it is a relatively fast-growing (generation time approximately 4 h) and a containment level 2 pathogen, M. marinum is used as a model organism to understand pathogenicity and host response to M. tuberculosis, which is slower growing (generation time approximately 20 h), and requires higher levels of containment[2]. The natural host, zebrafish, is used as a model organism to develop understanding of mycobacterial pathogenesis in general[3], but M. marinum has also been studied in mouse models of topical or systemic infections[4,5].
The primary host cell for M. tuberculosis infection is the macrophage where pathogenic mycobacteria initially infect and spread into resting macrophages. These in turn serve as a habitat for survival and replication of the bacteria[6]. Dissemination of mycobacteria by macrophages has been observed in real-time using M. marinum infected zebrafish embryos, showing the transfer of mycobacteria from one macrophage to another via membrane tethers[7]. Normally, activated macrophages possess potent microbicidal activity or at least control the growth of pathogenic mycobacteria[8]. However, the mycobacteria cell envelope confers to the bacilli the ability to survive and replicate within phagosomes of macrophages where they arrest phagosome maturation[9]. Macrophages respond to M. tuberculosis infection with apoptosis and secretion of inflammatory cytokines and are tightly involved in the formation of granulomas.
Mast cells have been found associated with tuberculous granuloma[10]. Abundant in mucosal tissues, intratracheal injection of guinea pigs with M. tuberculosis led to degranulation of mast cells[11]. Pre-treating mice with C48/80, a potent mast cell activator, by repeated intranasal administrations before intratracheal infection with M. tuberculosis impeded neutrophil and mononuclear cell recruitment in the bronchoalveolar space and clearance of bacteria[12]. This experiment illustrated the orchestrating role mast cells have in mounting a pro-inflammatory response. Reconstitution of mice with interleukin-3 (IL-3) differentiated bone marrow derived mast cells (BMDMC) from TLR2+/+ or TLR2-/- mice showed that TLR2 expression of mast cells was important for the normal inflammatory response to M. tuberculosis (myeloid cell recruitment, pro-inflammatory cytokine production, granuloma formation and bacterial clearance)[13].
Infection of the rat basophilic cell line RBL-2H3 (used as a model of mast cells) and peritoneal mast cells with M. tuberculosis triggered the release of pre-stored reactants such as histamine and β-hexosaminidase, and the de novo synthesis of pro-inflammatory cytokines TNF-α and IL-6 at 30 min and 6 h[14,15]. Mast cells may form cholesterol dependent pseudopod like structures (lipid rafts) to take up M. tuberculosis, which may then replicate within the mast cells[15].
However, it is unclear whether mycobacteria are susceptible to the inhibitory and/or killing actions posed by mast cells directly, whether and to what extent mast cells internalise mycobacteria. It is also not known to what extent mycobacteria survive inside the activated mast cell.
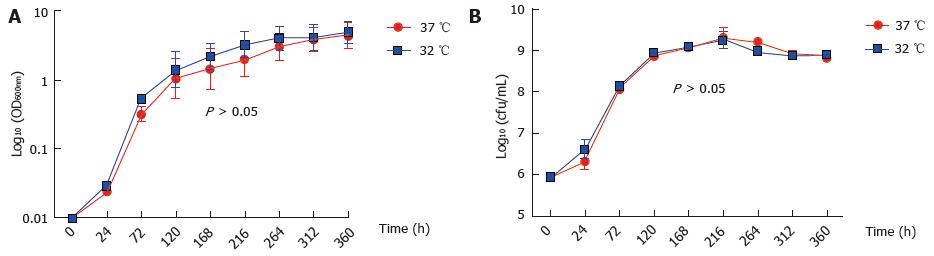
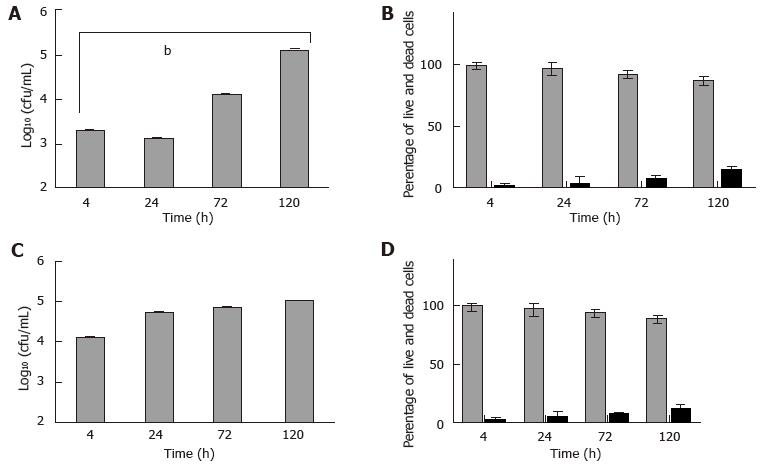
M. marinum BAA-535/M strain (isolated from human lesions) was originally obtained from the clinical laboratories of Moffitt Hospital at the University of California, United States, and provided by Dr. Hagedom M (University of Geneva). M. marinum strain expressing GFP was generated by introduction of pMind plasmid[16] expressing GFP from a Tet-promoter and was maintained in 50 μg/mL kanamycin. The bacilli were grown to mid log phase in Middlebrook 7H9 broth (Difco, Detroit, MI) supplemented with 0.5% (v/v) glycerol, 0.05% (v/v) Tween 80 and 10% (v/v) Albumin/Dextrose complex (ADC) at 32 °C without shaking. The strain was identified as M. marinum prior to experiments by acid fast staining, photochromogenic test, and sequencing of 16SrDNA. Single cell suspensions were obtained by passing through a 25G gauge needle to disperse cell clumps prior to infection. Whilst being a fish pathogen, the clinical isolate of M. marinum that was used in this work was able to grow at 37 °C (Figure 1).
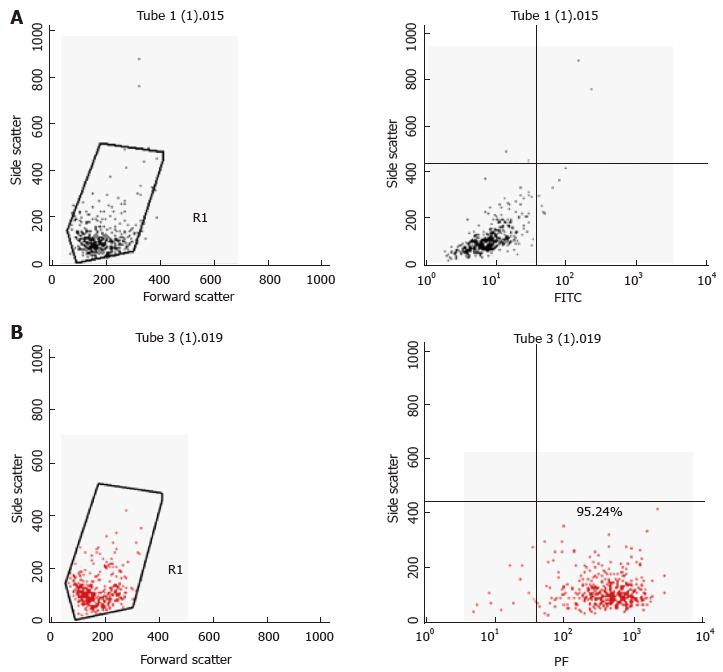
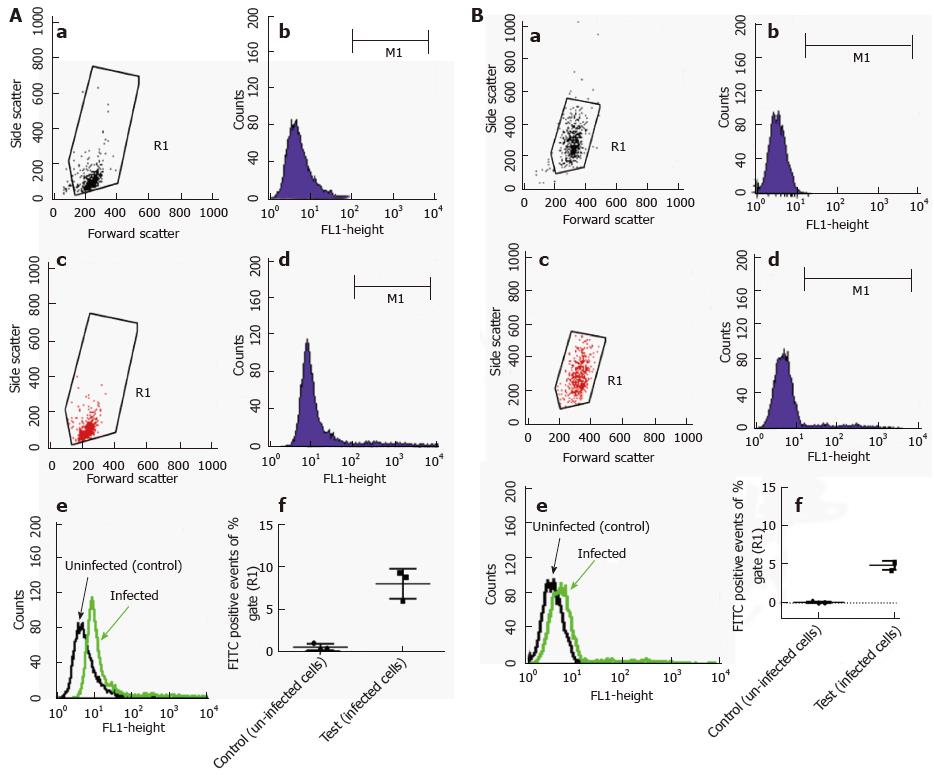
Primary murine mast cells were differentiated from bone marrows of mice (BMDMC) with 100 ng/μL of recombinant mouse IL-3 (Peprotech) over 5 wk[17]. Cells were characterised as mast cells by flow cytometry for the expression of high affinity mast cell receptor FceRI (BioLegend) (Figure 2). A human mast cell line (HMC-1) was cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% (v/v) heat-inactivated iron supplemented foetal calf serum (Fisher, Loughborough, United Kingdom), 1.2 mmol/L 1-thioglycerol (Sigma-Aldrich) and 100 U/mL penicillin, 100 μg/mL streptomycin. The cells were split 1:3 every 3-4 d and re-suspended in fresh medium. The human monocyte cell line MonoMac6[18] was maintained in RPMI1640 medium supplemented with 2 mmol/L L-glutamine, 1% (v/v) non-essential amino acids, 1 mmol/L pyruvate, 10% (v/v) heat-inactivated FCS and 100 U/mL penicillin, 100 μg/mL streptomycin.
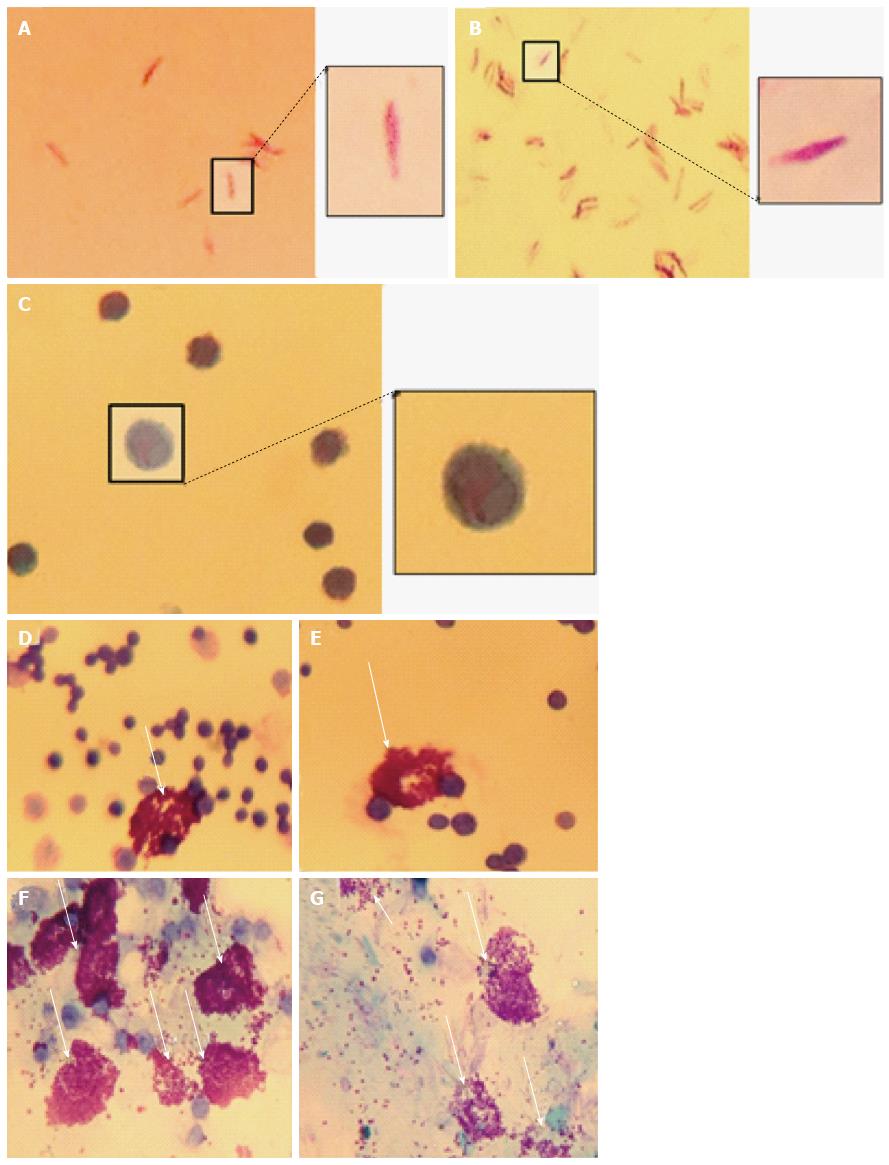
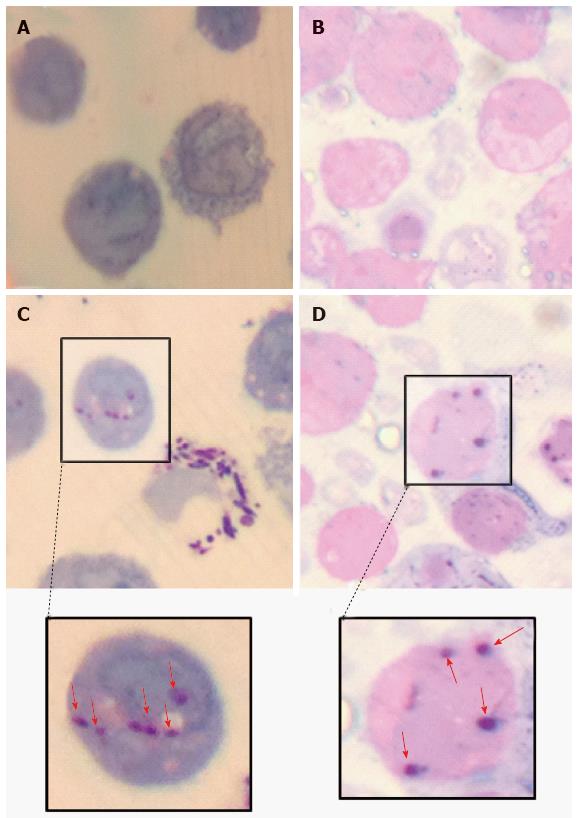
C57Bl/6J mice were used at 4 mo. A volume of 100 μL of heat-killed M. marinum (OD600nm 0.7), at a dose comparable to that of Complete Freund’s Adjuvant, was injected intraperitoneally in two mice, while two control mice received 100 μL PBS. All appropriate measures were taken to minimise pain or discomfort. The mice were monitored and culled at 4 h by cervical dislocation in the absence of any clinical signs of illness in any of the mice. Peritoneal lavage was performed with cold phosphate-buffered saline (PBS) with 4% (v/v) foetal calf serum (FCS) to collect peritoneal cells. Cytological staining of the peritoneal mast cells was then performed using Toluidine blue, Wright’s stain, Bismarck Brown and Kinyoun stain. Slides were air dried and mounted with cover slips using Xylene and DPX mounting medium.
Mast cells (2 × 105 cells/mL) were infected with M. marinum at a multiplicity of infection (MOI) of 0.5:1 (bacteria per cell) in 24 well-plates and incubated for 4 h (37 °C, 5% CO2 humidified atmosphere). Cells were washed three times with PBS and treated with 200 μg/mL amikacin for 2 h (optimised). Thereafter, the cells were washed and re-suspended with complete IMDM without antibiotics and incubated further for up to 120 h. At each time point, viability count (using trypan blue dye-exclusion method) and colony-forming units (CFU) count [after lysis of the infected cells with 0.1% (v/v) Triton X-100 for 5-10 min] were carried out. Ten microliters drops containing serially diluted bacteria were spotted on ADC supplemented 7H10 agar plates. The plates were stored in a plastic sleeve to maintain humidity and incubated at 32 °C for 7-14 d to obtain M. marinum colonies. For some experiments, cells (2 × 106 cells/mL) were infected with M. marinum at MOI 0.5, 10, 25, 50. At each time point cells were washed once with PBS to remove the extracellular bacteria and were processed for flow cytometry or stored at -80 °C for total RNA extraction. The supernatants were kept at -80 °C for further investigation.
The release of lactate dehydrogenase (LDH) from the cytoplasm in the supernatants was measured for BMDMCs infected with M. marinum at MOI 0.5, 10 and 25 for 8 h in comparison with uninfected control cells. The assay was performed according to the manufacturer’s instructions (CytoTox 96® Non-Radioactive Cytotoxicity Assay, Promega United Kingdom Ltd, Southampton, United Kingdom).
To assess purity of BMDMCs after differentiation with IL-3, the cell population was harvested (300 × g, 5 min); next, 1 × 106 cells were re-suspended with 100 μL Fc Block [CD16/32, clone 2.4G2, diluted 1:200 in FACS buffer, 2% (w/v) BSA in sterile PBS]. The samples were then incubated for 15 min on ice, centrifuged at 500 × g for 5 min at 4 °C and stained with 100 μL FACS buffer containing PE conjugated anti-mouse FcεRI alpha (clone MAR-1, BioLegend, 1:200) or its isotype control (Armenian Hamster IgG). The samples were incubated on ice for 15-30 min in the dark. After two-three washes in FACS buffer, the samples were re-suspended with 500 μL FACS buffer. The data were acquired using a FACSCalibur instrument (Becton Dickinson) and were analysed using the CellQuest Pro Software (Becton Dickinson) and Flow-Jo Software.
Quantification of mycobacterial uptake by mast cells was determined using an established fluorescence-quenching technique. A single cell suspension of M. marinum (1 × 109 CFU/mL) was labelled by incubation with 0.5 mg/mL of fluorescein isothiocyanate (FITC; Sigma) in 0.1 mol/L carbonate buffer (pH 9.0) at 37 °C for 2 h. FITC labelled M. marinum were washed twice with PBS to remove unbound FITC and re-suspended with fresh IMDM (complete without antibiotics). Thereafter, 1 × 106 cells/mL mast cells were infected at MOI 10:1 (bacteria per cell) for 24 h (37 °C in humidified 5% CO2 incubator). After each time point, the infected cells were washed three times with washing buffer [2% (w/v) BSA-PBS] to remove the extracellular bacteria and incubated with sodium acetate buffer (0.05 mol/L; pH 4.5) containing 0.06% trypan blue for 5 min at 4 °C. Cells were harvested (300 × g, 5 min) and re-suspended with 500 μL of fixation buffer [2% (w/v) paraformaldehyde in PBS] to carry out the acquisition step of the flow cytometer.
Discrimination of apoptotic and necrotic cellular subpopulations was achieved by simultaneously staining infected and uninfected HMC-1 cells, with Annexin V (FITC) and propidium iodide (PI) in the presence of 2.5 mmol/L of CaCl2. Samples were incubated on ice in the dark for 30 min and analysed by flow cytometry.
To visualise intracellular mycobacteria using GFP expressing M. marinum (pMIND), epifluorescence microscopy and confocal laser scanning microscope (Olympus FV1000) were applied. The cells were fixed with 4% (w/v) paraformaldehyde for 10 min at room temperature, followed by 30 min to dry in the dark. Thereafter, two drops of 3 μL mounting media [75% (v/v) glycerol] were added onto the cover slip and then pressed onto a microscope slide and allowed to adhere overnight at room temperature in the dark. For epifluorescence microscopy, the preparations were observed using a Nikon Diaphot 300 inverted microscope with a 100 W mercury light source. Images were recorded using a 12/10 bit, high speed peltier-cooled CCD camera (FDI, Photonic Science) using Image-Pro Plus (Media Cybernetics) software. Preparations for confocal laser scanning microscopy were observed using Olympus FV1000 inverted IX81 motorised confocal laser scanning microscope.
Zero point five micron-cross-sections of Mono Mac 6 and HMC-1 cells (infected with M. marinum at MOI 10 for 24 h) were produced using an ultramicrotome (Reichert, glass knife). After fixing the cells with 4% (v/v) glutaraldehyde in 0.1 mol/L phosphate buffer (pH 7.4), cells were post fixed with 1% (w/v) osmium tetroxide (OsO4) and dehydrated in an ethanol series. Cells were then exposed to the intermediate solvent epoxypropane (propylene oxide) before embedding in Agar Low Viscosity Resin (epoxy resin) and baking at 60 °C for 16 h. Sections were baked onto glass slides, then stained using Gram-Twort’s. All the steps were carried out on a 80 °C hotplate. The images were then viewed by light microscopy (Olympus CH2) using × 100 oil immersion and images were captured using iPhone 4S.
For transmission electron microscopy (TEM) analysis, cells (1 × 106 cells/mL) were infected with M. marinum at MOI 10 for 24, 72 and 120 h. After each timespan of infection, the cells were harvested (300 × g, 5 min) and washed once with PBS. The pellets were re-suspended with 1 mL 1% (v/v) glutaraldehyde in 0.1 mol/L sodium cacodylate buffer (pH 7.4) at RT for 30 min. After three washes of 0.1 mol/L sodium cacodylate at 250 × g for 20 min, the cells were fixed with 1% (w/v) osmium tetroxide in 0.1 mol/L sodium cacodylate for 90 min at 4 °C, and thoroughly washed with distilled de-ionised water three times for 20 min each wash[19]. The samples were embedded in 3% (w/v) agar and were dehydrated through an ethanol series. 100% analytical grade ethanol was used to wash the samples twice for 15 min, followed by two changes of propylene oxide (PO) for 15 min and then transferred through changes of PO: Agar low viscosity resin mix 1:1 ratio for 60 min in two changes and lastly 3:1 ratio for 60 min. The samples were transferred into 100% low viscosity agar overnight and then loaded into capsules with fresh agar low viscosity resin for 3 h, followed by polymerisation at 60 °C for 16 h to produce a solid block for ultrathin sectioning (80-100 nm, diamond knife).
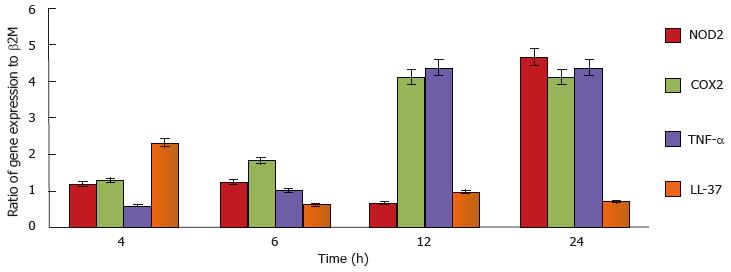
Total RNA was prepared using TRIzol Reagent (Invitrogen, Paisley, United Kingdom) according to the manufacturer’s instructions. After DNAseI digest and extraction, first strand cDNA synthesis was carried out according to the manufacturer’s instructions (Thermo Scientific United Kingdom). Primer sequences for subsequent PCR were for TNF-α 5’-CCCGACTATCTCGACTTTGC-3’ and 5’-GTTGGATGTTCGTCCTCCTC-3’, for NOD2 5’-GGCAGCCTCTTCAAAATGAG-3’ and 5’-GGGAAGAAGTCAATGGCAAA-3’, for COX2 5’-CGCCCTCATAATCATTTTCC-3’ and 5’-GAGGGCGATGAGGACTAGG-3’, for LL-37 5’-GAAGACCCAAAGGAATGGCC-3’ and 5’-CAGAGCCCAGAAGCCTGAGC-3’, for β2microglobulin 5’-GGCTATCCAGCGTACTCCAAAG-3’ and 5’-CAACTTCAATGTCGGATGGATG-3’. The Livak or 2-ΔΔCT method was used to calculate the normalised expression ratio of the target gene with reference to β2M gene[20].
Animal care and use statement: The animal protocol was designed to minimise pain or discomfort to the animals by using heat-killed M.marinum to cause immune stimulation, not disease. Mice were taken from a breeding colony within the unit, were acclimatised to 21 °C, 50% humidity, with 12/12 h light/dark cycle, and had ad libitum access to food (EURodent Diet 14%, LabDiet, International Product Supplies, London, United Kingdom) and water. Controls and experimental animals were each jointly housed. Intraperitoneal injection and cervical dislocation at the end of the experiment were performed by trained staff competent in these techniques in accordance with Home Office regulations.
Data are presented as means + standard deviations. Both Microsoft Excel and GraphPad Prism (v.4.02 for Windows; GraphPad Software, San Diego, CA) were used to analysis data in this study, and where appropriate statistical analyses was done using One-way and Two-way ANOVA (Analysis of Variance) with Bonferroni post-test using GraphPad Prism program, unless stated otherwise. A P value of < 0.05 was considered statistically significant.
To demonstrate mast cell involvement in an in vivo model, C57/Bl6J mice were stimulated intraperitoneally with heat-killed M. marinum and peritoneal lavages were performed. Cytologically, there was no evidence of bacteria associated to or visibly internalised by mast cells but significant degranulation (Figure 3). To assess the inflammatory response in a greater number of mast cells (replacement of animal models), a cell line was used. After incubation of HMC-1 with M. marinum for 4, 6, 12, 24 h, qRT-PCR analysis showed early expression of LL-37 mRNA, significant expression of COX-2 and TNF-α mRNA at 12 h and delayed expression of NOD2 mRNA at 24 h (Figure 4).
Assessment of LDH release, a measure of cytoplasmic leakage, from HMC-1 incubated with M. marinum revealed significant damage incrementally at MOI 10 and 25 compared to the uninfected control (Table 1). Flow cytometric analysis using Annexin V and PI showed that early apoptosis was detected in about 1/3 of the infected cell population. It is possible that internalised bacteria account for some PI staining in late apoptotic and necrotic cells (Table 2).
| Mast cells infected with M.marinumat different MOIs for 8 h | LDH release, % increase over uninfected control |
| 0.5:1 (bacteria:cell) | 4.20% |
| 10:1 (bacteria:cell) | 31.10% |
| 25:1 (bacteria:cell) | 43.70% |
| Uninfected/infectedHMC-1 | Annexin V-/PI- | Annexin V+/PI- | Annexin V+/PI+ | Annexin V-/PI+ |
| Uninfected (control) | 99.80% | 0.20% | 0% | 0% |
| Infected with M. marinum at MOI 10 | 58.52% | 31.76% | 2.98% | 6.70% |
| Infected with M. marinum at MOI 25 | 58.24% | 29.37% | 4.25% | 8.14% |
Next, HMC-1 and primary murine mast cells were infected with M. marinum (MOI 0.5) for up to 120 h. Among the aminoglycoside antibiotics tested, only amikacin proved efficacious against M. marinum, whilst leaving the viability of mast cells unimpaired (data not shown). It was used at 0.2 mg/mL for 2 h to kill all extracellular bacteria after infection of mast cells - this was verified by the lack of CFU grown from the supernatants - so that subsequently, the intracellular viable bacterial load could be determined. At 4 h after infection, there was significant presence of viable M. marinum in the experimentally lysed eukaryotic cells, especially in HMC-1. Cultures of HMC-1 and primary murine mast cells infected in parallel and analysed over longer times maintain a considerable intracellular bacterial load of M. marinum. Though intracellular growth of M. marinum appeared to be somewhat controlled in BMDMC at 24 h, both types of mast cells harboured significantly elevated CFU at 120 h with concomitant increase in cell damage (12%-14%), as assessed by the trypan blue exclusion method (Figure 5).
Flow cytometric analysis was performed using fluorescently labelled mycobacteria and quenching any extracellular signal with trypan blue. With this method, the intracellular content of the labelled material can be reliably measured. Because our GFP-expressing M. marinum strain showed growth restriction at 37 °C, it could not be used for this purpose. Alternatively, M. marinum were instead labelled with FITC and used to infect HMC1 and primary murine mast cells at MOI 10 for 24 h. After washing, trypan blue was added and cells were analysed by flow cytometry. Intracellular presence detected in this manner varied from 5%-8% of gated intact cells for BMDMC and HMC-1 cells, respectively (Figure 6).
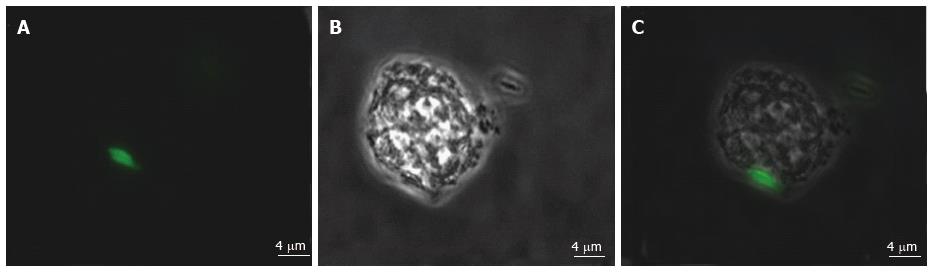
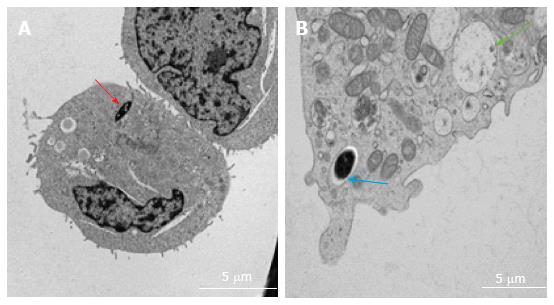
Various techniques were employed to visualise the intracellular presence of M. marinum in mast cells. Whilst fluorescence microscopy did not convincingly differentiate between adsorption and intracellular presence of M. marinum (Figure 7), clear evidence was for the first time obtained from the analysis of 0.5 μm cross-sections prepared of HMC-1 cells, in comparison with infected MonoMac6 cells (Figure 8). Though the monocytic cell line seems more avid in the bacterial uptake, a comparable picture is obtained when producing and analysing thin sections from mast cells. Lastly, to confirm this observation beyond doubt, TEM was used for both, HMC-1 cells and BMDMCs, which were infected with M. marinum at MOI 10 for 24 h (Figure 9). M. marinum sub-localised to the cytoplasm of HMC-1 cells whereas the bacilli sub-localised in vacuoles of BMDMCs.
This study shows that the involvement of mast cells in infection with M. marinum was manifold; mast cells degranulated, expressed mRNA of pro-inflammatory genes, lost cytoplasmic integrity, underwent apoptosis and to a lesser extent necrosis, and also internalised the bacilli. At the MOI chosen, the study could not evaluate a bactericidal or bacteriostatic effect of mast cell activity against mycobacteria, although the bacteria did not grow in culture medium alone.
We employed a human mast cell line (HMC-1 cells) which was established from a leukaemia patient by Butterfield et al[21]. As this cell line has a phenotype typical of immature mast cells, we isolated primary mast cells from the bone marrows of C57/Bl6J mice. The cells were differentiated with IL-3, and were 95% pure according to their surface staining for FcεRI. Other sources to study mast cell interaction with M. tuberculosis include a rat basophilic leukaemia cell line (RBL-2H3)[14,15] although there is contention about the relationship of these cells to mast cells[22].
Firstly, we established that M. marinum does lend itself to studies of eukaryotic infection because its growth at 37 °C compared well to that at the more typical temperature for this bacterium of 32 °C. This observation had precedence in the description by others of growth at 37 °C for the M. marinum strain[23-25]. In accordance with other studies, we found that GFP or ds-Red expressing M. marinum strains could not grow at 37 °C[26].
Next, we showed that M. marinum can infect, survive and replicate inside mast cells (HMC-1 and BMDMCs), where the bacilli sub-localise in the cytoplasm of HMC-1 cells, yet show a vacuolar presence in the more mature BMDMCs. In HMC-1 cells, no bacteria were seen surrounded by a vacuolar membrane at 24, 72 and 120 h (data not shown). Given that the overall uptake was low, it seems unlikely that M. marinum would have escaped from a vacuole in the interval of sampling time points. By contrast, M. marinum were regularly found in phagocytic vacuoles in the mature BMDMCs, with occasionally larger numbers of M. marinum occupying a single vacuole. These subcellular locations may reflect uptake mechanisms which differ between immature and mature, non-professional phagocytic cells.
During the analyses of the cytoplasmic presence of M. marinum in TEM images, the presence of damaged cells was noted. Viability of mast cells after low dose infection (MOI 0.5) for 120 h had shown only mild impairment. Measurement of cytoplasmic LDH release due to infection-induced injury revealed significant damage at MOI 10 and 25 compared to the uninfected control, contrary to low MOI (0.5). Up to this time point, it was known that M. marinum caused the release of LDH from infected macrophages[27]. In flow cytometric analysis using Annexin V and PI, we demonstrated that this damage was likely caused by an increase in necrotic cells.
Cruse et al[28] (2010) showed that mast cells (including HMC-1 cells) were able to kill Streptococcus pneumoniae and in response to pneumolysin, they rapidly released the antimicrobial peptide cathelicidin LL-37 (which is capable of inducing pneumococcal cell death), and the pro-inflammatory mediator LTC4. In relation to the pneumococcus, Cruse et al[28] (2010) also showed that although cell contact may increase the cytotoxicity of mast cells to pneumococci, HMC-1 cells can elicit antimicrobial activity to pneumococci in the absence of cell contact. In contrast, von Köckritz-Blickwede et al[29] (2008) demonstrated that HMC-1 cells can kill directly S. pyogenes without phagocytosis but cellular contact was required. They showed that LL-37 is caught in extracellular traps and H2O2 was required for extracellular trap formation (a process which is required for antimicrobial activity of HMC-1 against S. pyogenes). This process is at the cost of HMC-1 cell viability as the cells seemed to have undergone nuclear degradation to form these traps. The investigators referred to this so-called cell death, which is between apoptosis and necrosis, as “NETosis”. Whilst Arock et al[30] (1998) showed that mast cells phagocytose Staphylococcus aureus, Cruse et al[28] (2010) and von Köckritz-Blickwede et al[29] (2008) reported that mast cells do not phagocytose S. pneumoniae or S. pyogenes, we showed for the first time that mast cells can internalise M. marinum where the bacilli survive and replicate.
In spite of the fact that the viability of infected mast cells at MOI 0.5 decreases with prolonged infection (up to 120 h), a large proportion of the infected cells (HMC-1 and BMDMCs) were trypan blue negative and therewith deemed viable (88%-86%). Based on our analysis of intracellular viable count, it is these viable cells which harbour a significant number of viable M. marinum. These in turn are likely to have adapted to this intracellular environment and to the action of synthesised inflammatory products. mRNA for the antimicrobial peptide LL-37 was an early indicator of the antimycobacterial response. COX2 TNF-α and NOD2 mRNA were increased later. The release of TNF-α from the supernatants of infected HMC-1 cells at MOI 0.5, 1 and 10 for 24, 72 and 120 h, using TNF-α bio-assay (L929 cells) was undetectable (data not shown). This is consistent with previous observations that the TNF-α produced by HMC-1 is not bioactive[31]. Further studies are needed to evaluate the transcriptomic changes undergone by M. marinum in response to its intracellular localisation.
The Schlumberger Foundation Faculty for the Future is gratefully acknowledged for its continuous support throughout the fellowship (Sadiyo Siad). We thank Natalie Allcock and the late Stefan Hyman from the University of Leicester Electron Microscopy Unit. University of Leicester Biomedical Services staff is acknowledged for excellent maintenance and care of mouse colonies. We appreciate support from Professor Peter Andrew (University of Leicester) and helpful discussions with Dr Anthony Tsolaki (University of Brunel).
Host pathogen interactions in Mycobacterium tuberculosis (M. tuberculosis) infection have been extensively studied for macrophages, dendritic cells, and have recently been described for epithelial cells and mast cells. The study of M. tuberculosis, however, requires sophisticated containment facilities. Isolation of M. marinum from zebrafish, its characterisation in granulomatous waterborne disease in man and comparative analysis of its genome against other mycobacterial species eventually led to its wider introduction in experimental work. Mast cells are important immune effector cells, which tailor the release of their granular content to different modes of activation (crosslinking of FcεR bound antibodies, activation of complement, recognition of pathogen associated molecular patterns, integrin signalling). Whilst tissue resident macrophages originate from the yolk sac, mast cells differentiate from myeloid bone marrow precursors, are much longer lived than macrophages and proliferate. Mast cells reside close to mucosa, nerves, lymphatics and vessels.
M. smegmatis does not replicate intracellularly and incompletely models characteristics of M. tuberculosis. Use of M. marinum to study mammalian immune responses was hampered by its being primarily a pathogen of poikilotherms and causative of skin disease in humans (“fish tank granuloma”). However, its adaptation to 37 °C, its sensitivity to amikacin, and its ability to replicate intracellularly render M. marinum to be of interest to those studying host-pathogen interactions of M. tuberculosis.
This is the first study to use 0.5 μm thin cell sections to demonstrate the intracellular location of M. marinum after infection. This technique, for this question, rivals the use of transmission electron microscopy.
Because the natural host of M. marinum is different (zebra fish), the study does not address mycobacterial pathogenesis in the natural host. However, M. marinum does infect humans and causes fish tank granuloma which sometimes can resolve without treatment and implies an efficient, still not fully characterised role of the immune system. The authors do not know in which other ways the strain that has adapted to 37 °C differs from the isolate with an optimal temperature of 32 °C. The major focus of most recently published studies is on zebra fish, even so, fish tank granuloma is often misdiagnosed (probably, because of the authors’ lack of knowledge). Non-M. tuberculosis mycobacterial infections are an emerging clinical problem and they often require special drug regimes (different from M. tuberculosis). For these reasons, there is a need for an easy-to-handle, relevant, in vitro infection system. In addition, further studies could now evaluate the ability of M. marinum to adapt their metabolism and expression of lipid rich bodies in order to persist in cells, and thereby give an indication whether this is indeed a feature in those infected with M. marinum.
Primary mast cells describe those cells differentiated from mouse bone marrow precursors using Il-3 expressing FcεR.
The role of mast cells in infection secondary to M. marinum is explored in this study. The results demonstrate that a human mast cell line stimulated with M. marinum showed increased mRNA expression of pro-inflammatory genes including LL-37 and COX2/TNF-α, internalized the bacilli and underwent apoptosis. Furthermore the M. marinum can infect, survive and replicate inside mast cells.
| 1. | Rybniker J, Wolke M, Haefs C, Plum G. Transposition of Tn5367 in Mycobacterium marinum, using a conditionally recombinant mycobacteriophage. J Bacteriol. 2003;185:1745-1748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 2. | Cosma CL, Sherman DR, Ramakrishnan L. The secret lives of the pathogenic mycobacteria. Annu Rev Microbiol. 2003;57:641-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 279] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 3. | Prouty MG, Correa NE, Barker LP, Jagadeeswaran P, Klose KE. Zebrafish-Mycobacterium marinum model for mycobacterial pathogenesis. FEMS Microbiol Lett. 2003;225:177-182. [PubMed] |
| 4. | Watkins BY, Joshi SA, Ball DA, Leggett H, Park S, Kim J, Austin CD, Paler-Martinez A, Xu M, Downing KH. Mycobacterium marinum SecA2 promotes stable granulomas and induces tumor necrosis factor alpha in vivo. Infect Immun. 2012;80:3512-3520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Yamamoto Y, Saito H, Setogawa T, Tomioka H. Sex differences in host resistance to Mycobacterium marinum infection in mice. Infect Immun. 1991;59:4089-4096. [PubMed] |
| 6. | Wei J, Dahl JL, Moulder JW, Roberts EA, O’Gaora P, Young DB, Friedman RL. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J Bacteriol. 2000;182:377-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 116] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 7. | Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 410] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 8. | Mortaz E, Varahram M, Farnia P, Bahadori M, Masjedi MR. New Aspects in Immunopathology of Mycobacterium tuberculosis. ISRN Immunology. 2012;7:33-40. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 9. | Russell DG. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev. 2011;240:252-268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 232] [Cited by in RCA: 220] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 10. | Taweevisit M, Poumsuk U. High mast cell density associated with granulomatous formation in tuberculous lymphadenitis. Southeast Asian J Trop Med Public Health. 2007;38:115-119. [PubMed] |
| 11. | Ratnam S, Ratnam S, Puri BK, Chandrasekhar S. Mast cell response during the early phase of tuberculosis: an electron-microscopic study. Can J Microbiol. 1977;23:1245-1251. [PubMed] |
| 12. | Carlos D, de Souza Júnior DA, de Paula L, Jamur MC, Oliver C, Ramos SG, Silva CL, Faccioli LH. Mast cells modulate pulmonary acute inflammation and host defense in a murine model of tuberculosis. J Infect Dis. 2007;196:1361-1368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Carlos D, Frantz FG, Souza-Júnior DA, Jamur MC, Oliver C, Ramos SG, Quesniaux VF, Ryffel B, Silva CL, Bozza MT. TLR2-dependent mast cell activation contributes to the control of Mycobacterium tuberculosis infection. Microbes Infect. 2009;11:770-778. [PubMed] |
| 14. | Muñoz S, Hernández-Pando R, Abraham SN, Enciso JA. Mast cell activation by Mycobacterium tuberculosis: mediator release and role of CD48. J Immunol. 2003;170:5590-5596. [PubMed] |
| 15. | Muñoz S, Rivas-Santiago B, Enciso JA. Mycobacterium tuberculosis entry into mast cells through cholesterol-rich membrane microdomains. Scand J Immunol. 2009;70:256-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | Blokpoel MC, Murphy HN, O’Toole R, Wiles S, Runn ES, Stewart GR, Young DB, Robertson BD. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 2005;33:e22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 156] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 17. | Kalesnikoff J, Galli SJ. Antiinflammatory and immunosuppressive functions of mast cells. Methods Mol Biol. 2011;677:207-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 18. | Ziegler-Heitbrock HW, Thiel E, Fütterer A, Herzog V, Wirtz A, Riethmüller G. Establishment of a human cell line (Mono Mac 6) with characteristics of mature monocytes. Int J Cancer. 1988;41:456-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 439] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 19. | Minai-Fleminger Y, Elishmereni M, Vita F, Soranzo MR, Mankuta D, Zabucchi G, Levi-Schaffer F. Ultrastructural evidence for human mast cell-eosinophil interactions in vitro. Cell Tissue Res. 2010;341:405-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [PubMed] |
| 21. | Butterfield JH, Weiler D, Dewald G, Gleich GJ. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leuk Res. 1988;12:345-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 538] [Cited by in RCA: 582] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 22. | Stone KD, Prussin C, Metcalfe DD. IgE, mast cells, basophils, and eosinophils. J Allergy Clin Immunol. 2010;125:S73-S80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 977] [Cited by in RCA: 902] [Article Influence: 56.4] [Reference Citation Analysis (0)] |
| 23. | Clark HF, Shepard CC. Effect of environmental temperatures on infection with mycobacterium marinum (balnei) of mice and a number of poikilothermic species. J Bacteriol. 1963;86:1057-1069. [PubMed] |
| 24. | Bercovier H, Vincent V. Mycobacterial infections in domestic and wild animals due to Mycobacterium marinum, M. fortuitum, M. chelonae, M. porcinum, M. farcinogenes, M. smegmatis, M. scrofulaceum, M. xenopi, M. kansasii, M. simiae and M. genavense. Rev Sci Tech. 2001;20:265-290. [PubMed] |
| 25. | Kent ML, Watral V, Wu M, Bermudez LE. In vivo and in vitro growth of Mycobacterium marinum at homoeothermic temperatures. FEMS Microbiol Lett. 2006;257:69-75. [PubMed] |
| 26. | Ramakrishnan L. Using Mycobacterium marinum and its hosts to study tuberculosis. Curr Sci India. 2004;86:82-92. |
| 27. | Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, McDonald KL, Szyk A, LaRonde-LeBlanc N, Gao LY. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect Immun. 2008;76:5478-5487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 235] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 28. | Cruse G, Fernandes VE, de Salort J, Pankhania D, Marinas MS, Brewin H, Andrew PW, Bradding P, Kadioglu A. Human lung mast cells mediate pneumococcal cell death in response to activation by pneumolysin. J Immunol. 2010;184:7108-7115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 29. | von Köckritz-Blickwede M, Goldmann O, Thulin P, Heinemann K, Norrby-Teglund A, Rohde M, Medina E. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood. 2008;111:3070-3080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 423] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 30. | Arock M, Ross E, Lai-Kuen R, Averlant G, Gao Z, Abraham SN. Phagocytic and tumor necrosis factor alpha response of human mast cells following exposure to gram-negative and gram-positive bacteria. Infect Immun. 1998;66:6030-6034. [PubMed] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Lichtor T, Pani SP S- Editor: Ji FF L- Editor: A E- Editor: Wu HL