Published online Nov 26, 2015. doi: 10.4331/wjbc.v6.i4.409
Peer-review started: May 30, 2015
First decision: June 18, 2015
Revised: July 22, 2015
Accepted: September 10, 2015
Article in press: September 16, 2015
Published online: November 26, 2015
Processing time: 179 Days and 14.7 Hours
AIM: To identify non-quinazoline kinase inhibitors effective against drug resistant mutants of epidermal growth factor receptor (EGFR).
METHODS: A kinase inhibitor library was subjected to screening for specific inhibition pertaining to the in vitro kinase activation of EGFR with the gatekeeper mutation T790M, which is resistant to small molecular weight tyrosine kinase inhibitors (TKIs) for EGFR in non-small cell lung cancers (NSCLCs). This inhibitory effect was confirmed by measuring autophosphorylation of EGFR T790M/L858R in NCI-H1975 cells, an NSCLC cell line harboring the gatekeeper mutation. The effects of a candidate compound, Janus kinase 3 (JAK3) inhibitor VI, on cell proliferation were evaluated using the MTT assay and were compared between T790M-positive and -negative lung cancer cell lines. JAK3 inhibitor VI was modeled into the ATP-binding pocket of EGFR T790M/L858R. Potential physical interactions between the compound and kinase domains of wild-type (WT) or mutant EGFRs or JAK3 were estimated by calculating binding energy. The gatekeeper residues of EGFRs and JAKs were aligned to discuss the similarities among EGFR T790M and JAKs.
RESULTS: We found that JAK3 inhibitor VI, a known inhibitor for JAK3 tyrosine kinase, selectively inhibits EGFR T790M/L858R, but has weaker inhibitory effects on the WT EGFR in vitro. JAK3 inhibitor VI also specifically reduced autophosphorylation of EGFR T790M/L858R in NCI-H1975 cells upon EGF stimulation, but did not show the inhibitory effect on WT EGFR in A431 cells. Furthermore, JAK3 inhibitor VI suppressed the proliferation of NCI-H1975 cells, but showed limited inhibitory effects on the WT EGFR-expressing cell lines A431 and A549. A docking simulation between JAK3 inhibitor VI and the ATP-binding pocket of EGFR T790M/L858R predicted a potential binding status with hydrogen bonds. Estimated binding energy of JAK3 inhibitor VI to EGFR T790M/L858R was more stable than its binding energy to the WT EGFR. Amino acid sequence alignments revealed that the gatekeeper residues of JAK family kinases are methionine in WT, similar to EGFR T790M, suggesting that TKIs for JAKs may also be effective for EGFR T790M.
CONCLUSION: Our findings demonstrate that JAK3 inhibitor VI is a gatekeeper mutant selective TKI and offer a strategy to search for new EGFR T790M inhibitors.
Core tip: Non-small cell lung cancers caused by mutations in the epidermal growth factor receptor (EGFR) initially respond to tyrosine kinase inhibitors (TKIs). However, the therapeutic efficacy of EGFR-TKIs is limited by drug-resistant mutations such as the gatekeeper mutation T790M. Our present study rediscovered JAK3 inhibitor VI, a known TKI for Janus kinases (JAKs), as a selective EGFR T790M inhibitor. Our structural analysis revealed similarities among EGFR T790M and JAKs, offering a possible strategy to search for EGFR T790M inhibitors from known kinase inhibitors. Repositioning of the existing therapeutics may facilitate solving clinical problems such as drug resistance and toxicity.
- Citation: Nishiya N, Sakamoto Y, Oku Y, Nonaka T, Uehara Y. JAK3 inhibitor VI is a mutant specific inhibitor for epidermal growth factor receptor with the gatekeeper mutation T790M. World J Biol Chem 2015; 6(4): 409-418
- URL: https://www.wjgnet.com/1949-8454/full/v6/i4/409.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i4.409
Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) such as gefitinib and erlotinib are clinical therapeutics effective on non-small cell lung cancers (NSCLCs) with activating mutation in EGFR[1,2]. Most frequent activating mutations in EGFR, a point mutation L858R and an exon19 deletion (delE746-A750), increase affinity for EGFR-TKIs relative to that for wild type (WT)[3,4]. However, the development of drug-resistant mutations such as the gatekeeper mutation T790M limits clinical efficacies of the EGFR-TKIs and is the major cause of acquired resistance in NSCLC patients[5,6].
An irreversible EGFR-TKI with a reactive acceptor group covalently binds to a conserved cysteine residue in the EGFR kinase domain (Cys 797) via a Michael addition reaction. The covalent bond formation increases stability of the complex formation between a TKI and the ATP-binding site of the kinase domain in comparison with reversible inhibitors. Therefore, irreversible EGFR-TKIs provide the ability to inhibit EGFR T790M[7-12]. However, EGFR T790M inhibition by the quinazoline-based irreversible TKIs causes simultaneous inhibition of the WT EGFR because of similar ATP affinity between WT and the T790M mutant, resulting in adverse effects such as skin rash and diarrhea. Therefore, the use of irreversible EGFR-TKIs has been limited in gefitinib- or erlotinib-resistant NSCLCs and by the dose-limiting toxicity[13,14].
Here we report that Janus kinase 3 (JAK3) inhibitor VI[15], a known TKI for a tyrosine kinase JAK3, selectively inhibits EGFR T790M/L858R, but has less inhibitory effect on the WT EGFR in vitro. JAK3 inhibitor VI also suppresses the proliferation of NCI-H1975 cells, an NSCLC cell line harboring EGFR T790M/L858R. This inhibitor is modeled into the ATP-binding pocket of EGFR T790M/L858R and forms potential hydrogen bonds with the kinase domain of EGFR T790M/L858R. The gatekeeper residue of JAK3 is methionine in WT, and the overall structure of the catalytic domain of JAK3 is closely related to that of EGFR. Our findings indicate that a JAK3 inhibitor is a mutant-selective reversible TKI for EGFR T790M.
JAK3 inhibitor VI and JAK inhibitor I were purchased from Calbiochem (San Diego, CA). Gefitinib was purchased from JS Research Chemical Trading (Wedel, Germany). SCADS inhibitor kit 3 was provided by Screening Committee of Anticancer Drugs supported by a Grant-in-Aid for Scientific Research on Priority Area “Cancer” from The Ministry of Education, Culture, Sports, Science and Technology, Japan. Antibodies against EGFR, pEGFR (Y1045), pEGFR (Y1068), and STAT3 were purchased from Cell Signaling Technology (Beverly, MA). Anti-pSTAT3 (Y705) was purchased from BD (Franklin Lakes, NJ). HRP-conjugated anti-phosphotyrosine antibodies PY20 and 4G10 were purchased from BD and Millipore (Billerica, MA). NCI-H1975 cell line was obtained from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium supplemented with 2 mmol/L L-glutamine and 10% Fetal Bovine Serum at 37 °C in a humidified atmosphere of 5% CO2.
One hundred nanograms of recombinant cytoplasmic domains (amino acid residues 696 to the C-terminus) of the WT EGFR, L858R, or T790M/L858R (Cell Signaling Technology) were preincubated with kinase inhibitors in 25 μL of kinase reaction buffer (120 mmol/L HEPES, pH 7.5; 10 mmol/L MgCl2; 10 mmol/L MnCl2; 6 μmol/L Na3VO4; and 2.5 mmol/L DTT) at 25 °C for 30 min. Subsequently, 25 μL of ATP/substrate solution containing 2 or 6 μmol/L ATP and 6 μmol/L poly (Glu-Tyr) biotinylated peptide (Cell Signaling Technology) was added to the preincubation mixture. The kinase reaction was performed at 25 °C for 30 min and terminated by adding 50 μL of stop buffer (50 mmol/L EDTA, pH 8.0). Kinase activity was estimated using ELISA with avidin-coated 96-well plates and anti-phosphotyrosine antibodies (PY20 and 4G10).
NCI-H1975 or A431 cells were pretreated using JAK3 inhibitor VI for 1 h in 0.2% serum conditions and were stimulated using 200 ng/mL EGF for 15 min. Alternatively, NCI-H1975 cells were treated with kinase inhibitors at the indicated concentration for 16 h. Cells were lysed in RIPA buffer (50 mmol/L Tris-HCl, pH 7.4; 150 mmol/L NaCl; 1% Triton X-100; 1% sodium deoxycholate; 0.1% SDS; 1 mmol/L NaF; 1 mmol/L Na3VO4; and protease inhibitors) and cleared by centrifugation. SDS sample buffer was added to the supernatants. Protein samples were separated using SDS-PAGE and analyzed using Western blotting with antibodies against phospho-specific or total proteins.
Proliferation assay
Cells were cultivated in a flat-bottomed 96-well plate at 1 × 104 cells per well in 150 μL media containing various concentrations of kinase inhibitors for 72 h. The inhibitor-treated cells were incubated in the presence of 0.5 mg/mL 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) in a CO2 incubator for 4 h. One hundred microliters of 20% SDS was added to each well for dissolving the insoluble purple formazan product into a colored solution. The absorption at 570 nm was measured using a spectrophotometer. The means of data values from at least three independent experiments were calculated.
All starting templates, except the WT EGFR, were prepared by superimposing the crystal structure of the WT EGFR (PDB ID: 4WKQ) onto crystal structures of EGFR-LRTM (PDB ID: 4I22) and JAK3 (PDB ID: 3LXL) without ligands. Docking studies were performed to analyze interactions of some inhibitors in the binding site of the WT EGFR, EGFR-TMLR, and JAK3 through PyRx[16] using AutoDock 4.2.6[17]. The adopted standard protein preparation protocol involves the addition of missing hydrogen atoms to the starting template and the assignment of an ionizable state to each charged group. The docking grid of 11 Å × 12 Å × 14 Å was generated based on the observed binding site of the WT EGFR using gefitinib (PDB ID: 4WKQ). Schematic representations were generated using the UCSF Chimera package[18].
All data are presented as the mean ± SE. The Mann–Whitney U-test was used to determine statistically significant differences.
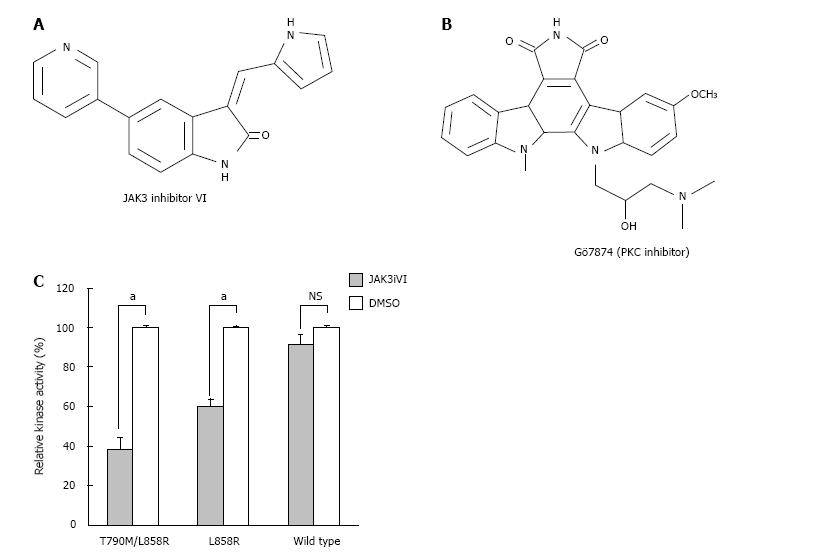
To profile sensitivities of EGFR mutants against different classes of kinase inhibitors, we assayed in vitro recombinant cytoplasmic domains of the WT EGFR, EGFR L858R, or EGFR T790M/L858R for 95 kinase inhibitors (Table 1). Several compounds such as Ellagic acid, Damnacanthal, and RAF1 kinase inhibitor I inhibited all three EGFR kinases activity, but other kinase inhibitors showed selectivity in their inhibitory activity against WT or mutant kinases. Among inhibitors with selectivity, JAK3 inhibitor VI[15] and Gö7874[19] reduced kinase activity of EGFR T790M/L858R. Gö7874 is a known PKC inhibitor and has an indolocarbazole structure that is also seen in staurosporine, a classical kinase inhibitor (Figure 1B). Because it has been reported that indolocarbazole compounds selectively inhibit EGFR T790M but show limited effects on the WT EGFR[20], JAK3 inhibitor VI was used in further studies. JAK3 inhibitor VI, a 3′-pyridyl oxindole compound (Figure 1A), inhibited EGFR T790M/L858R (IC50 = 1.43 μmol/L) mutant, but weaker effects were observed on the L858R mutant (IC50 > 10 μmol/L) and WT EGFR (IC50 > 10 μmol/L) (Figure 1C).
| Target | Compound name |
| AK | ABT-702 |
| AKT (PKB) | Akt Inhibitor IV, Akt Inhibitor VIII, Akt Inhibitor XI |
| AMPK | compound C |
| ATM | ATM/ATR kinase inhibitor, ATM kinase inhibitor |
| Aurora kinase | Aurora kinase/CDK inhibitor, aurora kinase inhibitor II, Aurora kinase inhibitor III |
| Bcr-abl | AG957 |
| BTK | LFM-A13, terreic acid |
| CAMKII | KN-93, KN-62, Lavendustin C |
| CDK | Kenpaullone, purvalanol A, Olomoucine, Alsterpaullone, Cdk1/2 inhibitor III, Cdk2/9 inhibitor, NU6102, Cdk4 inhibitor, NSC625987 |
| Chk | SB218078, Isogranulatimide, Chk2 inhibitor, Chk2 inhibitor II |
| CK | Ellagic acid (dihydrate), TBB, DMAT, D4476 |
| Clk | TG003 |
| DGK | Diacylglycerol kinase inhibitor II |
| DNA-PK | IC60211 |
| eEF2 kinase | TX-1918 |
| EGFR | BPIQ-II, AG1478, AG490 |
| FGFR | SU4984, SU5402 |
| Flt-3 | Flt-3 Inhibitor |
| Fms | cFMS Receptor Inhibitor |
| Fyn | SU6656 |
| GSK | GSK-3 inhibitor IX, 1-Azakenpaullone, Indirubin-3’-monoxime |
| HER2 | AG825 |
| IGF-IR | AG1024, AGL 2263 |
| IKK | BMS-345541, IKK-2 inhibitor VI |
| IRAK | IRAK-1/4 inhibitor |
| JAK | JAK Inhibitor I, JAK3 Inhibitor VI |
| JNK | SP600125, JNK inhibitor VIII |
| Lck | Damnacanthal, PP2 |
| MAPK | ERK inhibitor II |
| MEK | PD98059, U-0126, MEK inhibitor I |
| Met | SU11274 |
| MLCK | ML-7 |
| p38 MAPK | SB202190, SB239063 |
| PDGFR | AG1296, SU11652, PDGF receptor inhibitor V, PDGF receptor inhibitor IV |
| PI3K | LY-294002, Wortmannin |
| PKA | H-89, 4-cyano-3-methylisoquinoline |
| PKC | Bisindolymaleimide I, Go7874 |
| PKG | Rp-8-CPT-cGMPS, KT5823 |
| PKR | PKR inhibitor |
| Raf | RAF1 kinase inhibitor I, ZM 336372 |
| ROCK | H-1152, Y-27632 |
| Hsp90 | radicicol |
| Src | PP1 analog |
| Syk | Syk inhibitor |
| TGF-bRI | SB431542, TGF-beta RI kinase inhibitor II |
| Tpl2 | Tpl2 kinase inhibitor |
| TrKA | TrkA inhibitor |
| VEGFR | VEGFR receptor inhibitor II, VEGF recptor 2 inhibitor I, SU1498 |
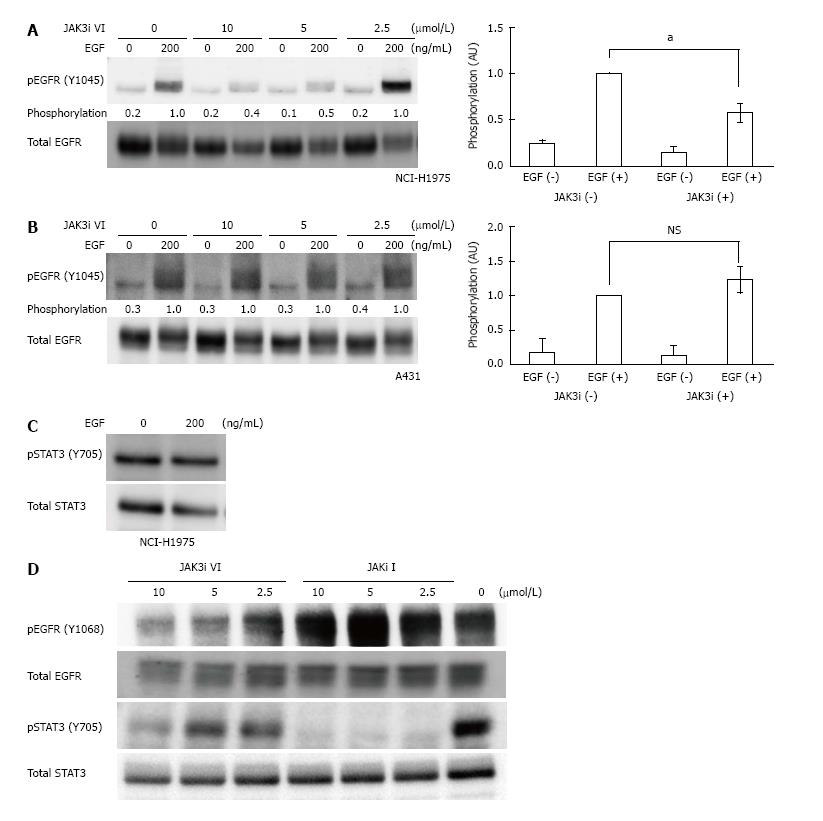
Next, we tested whether JAK3 inhibitor VI decreased EGFR signaling in the EGFR T790M/L858R-positive NCI-H1975 cells. EGF-induced EGFR autophosphorylation at Y1045 was significantly reduced in 10 or 5 μmol/L JAK3 inhibitor VI-treated NCI-H1975 cells (Figure 2A). In contrast, JAK3 inhibitor VI had no effect on the WT EGFR Y1045 phosphorylation in A431 cells (Figure 2B). These data confirm the mutant EGFR-specific blockage by JAK3 inhibitor VI in the T790M/L858R positive NCI-H1975 cells.
Although JAK 3 inhibitor VI specifically inhibited EGFR T790M/L858R in cells, it was not clear whether the T790M/L858R inhibition was a direct effect on the EGFR kinase or an indirect effect via JAK3 inhibition. To clarify this point, we analyzed phosphorylation levels of STAT3, a direct JAK3 substrate. The STAT3 phosphorylation level was unchanged even after stimulation using 200 ng/mL EGF, which efficiently elevated EGFR Y1045 phosphorylation (Figure 2A and C). In addition to the effects of JAK3 inhibitor VI on the EGF-stimulated phosphorylation of EGFR T790M/L858R, those on the basal phosphorylation of EGFR T790M/L858R and STAT3 were analyzed pertaining to NCI-H1975 cells in a 10% serum culture condition. In contrast to a decrease in the phosphorylation level of EGFR T790M/L858R at 5 μmol/L JAK3 inhibitor VI, STAT3 Y705 phosphorylation was apparent at 5 μmol/L and began to decrease from a higher concentration, 10 μmol/L (Figure 2D). Another JAK inhibitor, JAK inhibitor I, significantly reduced STAT3 Y705 phosphorylation at 2.5 μmol/L, but did not affect EGFR Y1068 phosphorylation at higher concentrations such as 5 or 10 μmol/L (Figure 2D). These data suggest that JAK3 inhibitor VI reduces the phosphorylation level of EGFR T790M/L858R without JAK3 inhibition, and that JAKs are not major contributors to EGFR phosphorylation in NCI-H1975 cells.
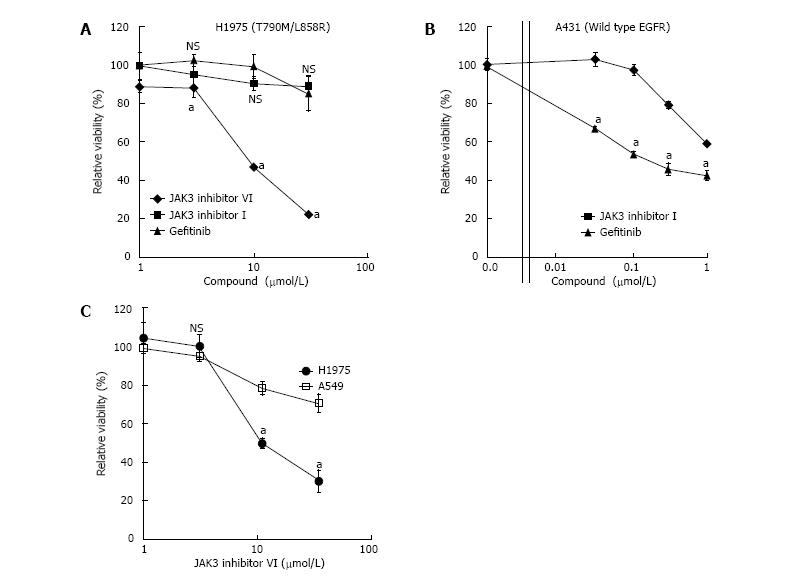
Next, we performed MTT assays to test the effects of JAK3 inhibitor VI on proliferation of EGFR T790M/L858R positive NCI-H1975 cells. NCI-H1975 cells were cultured in 96-well plates for 3 d with or without different concentrations of compounds. JAK3 inhibitor VI showed growth suppression at 10 and 30 μmol/L, but gefitinib or JAK inhibitor I did not (Figure 3A). The effects of JAK3 inhibitor VI on proliferation of the WT EGFR expressing A431 (Figure 3B) and A549 (Figure 3C) cells were also analyzed. These cells were less sensitive to JAK3 inhibitor VI than NCI-H1975 cells.
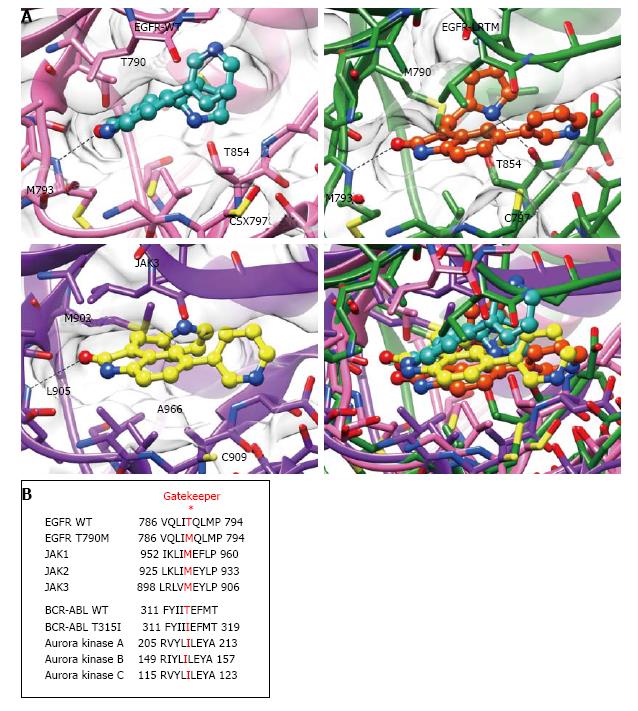
To obtain structural insight, docking simulations between JAK3 inhibitor VI and kinases were performed. In the model structure, JAK3 inhibitor VI bound to the ATP-binding pocket of EGFR T790M/L858R and formed hydrogen bonds with M793 and T854 residues (Figure 4A). Estimated binding to EGFR T790M/L858R was more stable than that to the WT EGFR and was equivalent with that to JAK3 (EGFR T790M/L858R: -7.38 kcal/mol; JAK3: -7.59 kcal/mol; WT EGFR: -6.23 kcal/mol). Amino acid sequence alignments of the EGFR T790 flanking regions and the corresponding regions of JAKs revealed that the gatekeeper residues of WT JAKs are methionine, similar to those of EGFR T790M (Figure 4B).
EGFR T790M limits the clinical efficacy of EGFR-TKIs. Irreversible TKIs for EGFR T790M are associated with toxic effects due to simultaneous inhibition of the WT EGFR. In the present study, we found that JAK3 inhibitor VI selectively inhibits EGFR T790M/L858R and had a lesser inhibitory effect on the WT EGFR in vitro. JAK3 inhibitor VI also suppressed the proliferation of NCI-H1975 cells harboring EGFR T790M/L858R. Thus, our findings demonstrate that a JAK3 inhibitor is a mutant selective TKI for EGFR T790M/L858R and offer a potential strategy to develop EGFR T790M-specific inhibitors by applying known inhibitors for JAK family kinases closely related to the ErbB family.
JAK3 inhibitor VI selectively inhibits the gefitinib-resistant EGFR T790ML858R. JAK3 inhibitor VI (10 μmol/L) inhibited kinase activity of EGFR T790M/L858R; however, it was ineffective on the activity of the WT EGFR in vitro kinase assay (Figure 1C). Furthermore, JAK3 inhibitor VI efficiently reduced autophosphorylation of EGFR T790M/L858R in gefitinib-resistant lung cancer cells, NCI-H1975 (Figure 2A), in contrast to resistance of the WT EGFR in A431 cells (Figure 2B). Dosage restriction of quinazoline-based EGFR-TKI treatments is attributable to the toxicity derived from simultaneous inhibition of the WT EGFR. As a possible solution for the issue, Zhou et al[21] reported that compounds with a non-quinazoline structure, pyrimidine, were specifically effective on mutant EGFRs. Staurosporine-related indolocarbazole compounds such as Gö6976 and PKC412 are also more selective for EGFR T790M than quinazoline-based EGFR inhibitors[19]. Therefore, applications of non-quinazoline compounds can confer T790M mutant specific inhibition without affecting the WT EGFR. Our data indicate that JAK3 inhibitor VI, an indol-based compound, provides an additional example of EGFR T790M-specific structures.
JAK3 inhibitor VI suppresses proliferation of NSCLCs harboring the gatekeeper mutation EGFR T790M. EGFR T790M/L858R-expressing NCI-H1975 cells showed reduced proliferation in the presence of JAK3 inhibitor VI, but not in the presence of gefitinib or JAK inhibitor I (Figure 3A). Furthermore, WT EGFR-expressing cell lines, A431 and A549, were less sensitive to JAK3 inhibitor VI in comparison with NCI-H1975 cells (Figures 3B and C), suggesting specific inhibition of EGFR T790M by JAK3 inhibitor VI. Because JAK3 inhibitors block JAK3 that may phosphorylate EGFR, indirect effects via JAK3 inhibition may cause a reduction in autophosphorylation of EGFR T790M/L858R in NCI-H1975 cells. However, EGFR T790M/L858R inhibition was observed in lower concentrations of JAK3 inhibitor VI than those of JAK3 inhibition. Another JAK inhibitor, JAK inhibitor I, did not show EGFR T790M/L858R inhibition. Furthermore, T790M kinase inhibition was observed in an in vitro assay using recombinant kinases. Therefore, the decrease in autophosphorylation of EGFR T790M/L858R seems to be caused by direct inhibition in EGFR T790M/L858R rather than by JAK3 inhibition-mediated indirect effects.
Structural characteristics of EGFR T790M mutants may be analogous to those of JAK3. In the docking study, JAK3 inhibitor VI bound to the ATP-binding pocket of EGFR T790M/L858R with an additional hydrogen bond (Figure 4A). Binding energy calculations predicted that JAK3 inhibitor VI would more stably bind to EGFR T790M/L858R than to the WT EGFR with an equivalent stability to the simulated binding to JAK3 (Figure 4A). Furthermore, EGFR and JAK family kinases are located in neighboring clans in the human kinome[22], indicating that overall structures of the catalytic domains of JAKs are closely related to those of EGFRs. The gatekeeper residues of WT JAKs are methionines, which are identical to the gatekeeper mutant EGFR T790M (Figure 4B). Sequence alignments of the gatekeeper flanking regions between BCR-ABLs and Aurora kinases indicated that the corresponding residues in Aurora kinases are isoleucines, similar to the gatekeeper mutation T315I. These findings suggest a hypothesis in which inhibitor sensitive kinases such as EGFR L858R, exon19-deleted EGFR, and BCR-ABL obtained structural similarities to other kinases during acquisition of drug resistance. In fact, Aurora kinase inhibitors are effective on BCR-ABL T315I[23,24] in addition to our findings of JAK3 inhibitor VI as an EGFR T790M inhibitor. Therefore, it may be a rational strategy to screen compounds effective on different kinases structurally similar to gatekeeper mutants in the fight against drug resistant kinases.
We thank the Screening Committee of Anticancer Drugs in the Scientific Support Programs for Cancer Research Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology for SCADS inhibitor kits and Chie Ishikawa and Emi Takahashi for technical assistance.
Tyrosine kinase inhibitors against the epidermal growth factor receptor (EGFR-TKIs) are initially effective on non-small cell lung cancers (NACLCs) caused by EGFR mutations. However, drug-resistant mutations such as the gatekeeper mutation T790M limit the clinical efficacy of EGFR-TKIs.
Irreversible EGFR-TKIs that covalently bind to the EGFR kinase domain have provided a way to inhibit the gatekeeper mutant EGFR T790M.
EGFR T790M inhibition by the quinazoline-based irreversible TKIs causes simultaneous inhibition of the WT EGFR, resulting in adverse effects such as skin rash and diarrhea. The present study rediscovered Janus kinases 3 (JAK3) inhibitor VI as a reversible EGFR-TKI selective for EGFR T790M.
The authors’ structural analysis revealed similarities among EGFR T790M and Janus kinases (JAKs). This may offer a new strategy to screen for EGFR T790M inhibitors from known kinase inhibitors.
JAKs are protein tyrosine kinases, whose catalytic domain structures are closely related to those of EGFRs.
In current manuscript entitled “JAK3 inhibitor VI is a mutant specific inhibitor for EGFR with gatekeeper mutation T790M”, the authors demonstrated that JAK3 inhibitor VI could specifically inhibit EGFR gatekeeper mutation (T790M) in non-small cell lung cancers.
| 1. | Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947-957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5906] [Cited by in RCA: 6600] [Article Influence: 388.2] [Reference Citation Analysis (21)] |
| 2. | Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1687] [Cited by in RCA: 1854] [Article Influence: 109.1] [Reference Citation Analysis (0)] |
| 3. | Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 904] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 4. | Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N, Sliwkowski MX. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163-8171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 345] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 5. | Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2470] [Cited by in RCA: 2706] [Article Influence: 128.9] [Reference Citation Analysis (0)] |
| 6. | Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3081] [Cited by in RCA: 3244] [Article Influence: 154.5] [Reference Citation Analysis (0)] |
| 7. | Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, Liang MC, Perera SA, Zaghlul S, Borgman CL. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 187] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 8. | Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, Zhao F, Vincent PW, Naumov GN, Bradner JE. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924-11932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 528] [Cited by in RCA: 607] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 9. | Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, Padera RF, Shapiro GI, Baum A, Himmelsbach F. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702-4711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1188] [Cited by in RCA: 1189] [Article Influence: 66.1] [Reference Citation Analysis (0)] |
| 10. | Miller VA, Hirsh V, Cadranel J, Chen YM, Park K, Kim SW, Zhou C, Su WC, Wang M, Sun Y. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. Lancet Oncol. 2012;13:528-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 699] [Cited by in RCA: 791] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 11. | Jänne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, Ahn MJ, Kim SW, Su WC, Horn L. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689-1699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1435] [Cited by in RCA: 1707] [Article Influence: 155.2] [Reference Citation Analysis (0)] |
| 12. | Jiang T, Zhou C. Clinical activity of the mutant-selective EGFR inhibitor AZD9291 in patients with EGFR inhibitor-resistant non-small cell lung cancer. Transl Lung Cancer Res. 2014;3:370-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 13. | Ramalingam SS, Blackhall F, Krzakowski M, Barrios CH, Park K, Bover I, Seog Heo D, Rosell R, Talbot DC, Frank R. Randomized phase II study of dacomitinib (PF-00299804), an irreversible pan-human epidermal growth factor receptor inhibitor, versus erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2012;30:3337-3344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 212] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 14. | Sequist LV, Besse B, Lynch TJ, Miller VA, Wong KK, Gitlitz B, Eaton K, Zacharchuk C, Freyman A, Powell C. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:3076-3083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 354] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 15. | Adams C, Aldous DJ, Amendola S, Bamborough P, Bright C, Crowe S, Eastwood P, Fenton G, Foster M, Harrison TK. Mapping the kinase domain of Janus Kinase 3. Bioorg Med Chem Lett. 2003;13:3105-3110. [PubMed] [DOI] [Full Text] |
| 16. | Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 845] [Cited by in RCA: 1895] [Article Influence: 172.3] [Reference Citation Analysis (0)] |
| 17. | Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785-2791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19127] [Cited by in RCA: 17439] [Article Influence: 1025.8] [Reference Citation Analysis (0)] |
| 18. | Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605-1612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42909] [Cited by in RCA: 35939] [Article Influence: 1633.6] [Reference Citation Analysis (0)] |
| 19. | Kleinschroth J, Hartenstein J, Rudolph C, Schächtele C. Novel indolocarbazole protein kinase c inhibitors with improved biochemical and physicochemical properties. Bioorg Med Chem Lett. 1995;5:55-60. [RCA] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Lee HJ, Schaefer G, Heffron TP, Shao L, Ye X, Sideris S, Malek S, Chan E, Merchant M, La H. Noncovalent wild-type-sparing inhibitors of EGFR T790M. Cancer Discov. 2013;3:168-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 21. | Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070-1074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 731] [Cited by in RCA: 823] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 22. | Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912-1934. [PubMed] |
| 23. | Young MA, Shah NP, Chao LH, Seeliger M, Milanov ZV, Biggs WH, Treiber DK, Patel HK, Zarrinkar PP, Lockhart DJ. Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680. Cancer Res. 2006;66:1007-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 209] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 24. | Modugno M, Casale E, Soncini C, Rosettani P, Colombo R, Lupi R, Rusconi L, Fancelli D, Carpinelli P, Cameron AD. Crystal structure of the T315I Abl mutant in complex with the aurora kinases inhibitor PHA-739358. Cancer Res. 2007;67:7987-7990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 103] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
P- Reviewer: Rangel-Corona R, Zaravinos A, Zhang L S- Editor: Tian YL L- Editor: A E- Editor: Lu YJ
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/