Published online May 26, 2014. doi: 10.4331/wjbc.v5.i2.216
Revised: January 15, 2014
Accepted: March 17, 2014
Published online: May 26, 2014
Processing time: 208 Days and 17.4 Hours
Krüppel-like factor (KLF) family proteins are transcription factors that regulate numerous cellular functions, such as cell proliferation, differentiation, and cell death. Posttranslational modification of KLF proteins is important for their transcriptional activities and biological functions. One KLF family member with important roles in cell proliferation and tumorigenesis is KLF5. The function of KLF5 is tightly controlled by post-translational modifications, including SUMOylation, phosphorylation, and ubiquitination. Recent studies from our lab and others’ have demonstrated that the tumor suppressor FBW7 is an essential E3 ubiquitin ligase that targets KLF5 for ubiquitination and degradation. KLF5 contains functional Cdc4 phospho-degrons (CPDs), which are required for its interaction with FBW7. Mutation of CPDs in KLF5 blocks the ubiquitination and degradation of KLF5 by FBW7. The protein kinase Glycogen synthase kinase 3β is involved in the phosphorylation of KLF5 CPDs. In both cancer cell lines and mouse models, it has been shown that FBW7 regulates the expression of KLF5 target genes through the modulation of KLF5 stability. In this review, we summarize the current progress on delineating FBW7-mediated KLF5 ubiquitination and degradation.
Core tip: The protein levels of Krüppel-like factor (KLF)5 are tightly controlled in cell. Ubiquitination and destruction of KLF5 via FBW7, a famous tumor suppressor, has proved to have important roles in multiple cellular progresses by different studies. Here, we summarize these studies and show the physiological and pathological significance of FBW7-mediated degradation of KLF5.
- Citation: Luan Y, Wang P. FBW7-mediated ubiquitination and degradation of KLF5. World J Biol Chem 2014; 5(2): 216-223
- URL: https://www.wjgnet.com/1949-8454/full/v5/i2/216.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v5.i2.216
Krüppel-like factor (KLF) family proteins are important transcription factors that regulate numerous cellular processes[1]. KLF5 is a member of the KLF family that has been well-studied and shown to play a key role in mediating multiple cellular activities, such as proliferation and differentiation, in both normal and tumor cells[2]. Post-translational modifications of KLF5, including ubiquitination, SUMOylation, acetylation, and phosphorylation, can impact both the stability and activity of KLF5, thus affecting its downstream cellular functions[3-8].
FBW7 is the mammalian homolog of CDC4 in Saccharomyces cerevisiae and SEL10 in C. elegans. It is a component of the SCF (SKP1-CUL1-F-box protein) ubiquitin ligase complex. FBW7 is thought to have an important role in tumor biology by serving as a critical regulator of several oncoproteins, and mutations of FBW7 are found in a rapidly expanding number of human neoplasms[9].
In this review, we summarize the progress of research on FBW7-mediated KLF5 degradation and ubiquitination and show the physiological and pathological significance of KLF5 regulation by FBW7.
KLFs are a family of transcription factors with homologies to the Krüppel protein and the transcription factor Sp1 in Drosophila melanogaster and mammals, respectively[1]. To date, 17 mammalian KLFs have been identified, all of which contain three zinc finger motifs at the carboxyl-terminals, which are responsible for binding to GC-rich DNA sequences[10,11]. The KLFs have been demonstrated to play essential roles in development, immunity and cancer[1,10-15].
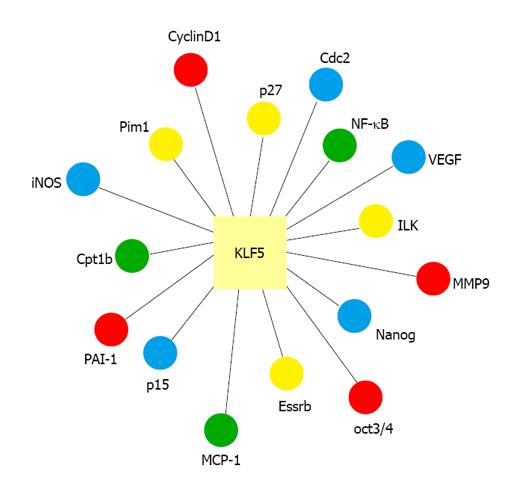
KLF5, also known as BTEB2 and IKLF, is an important KLF factor. KLF5 is widely expressed in various tissues, including lung, colon, intestine, and pancreas[2,16-19]. KLF5 is located at chromosomal position 13q22.1 in the human genome. It is involved in the regulation of diverse cellular functions, including cell cycle, proliferation, apoptosis, differentiation and stem cell self-renewal, by regulating the expression of numerous genes (Figure 1)[2,20-23]. Previous studies have shown that KLF5 plays a pivotal role in regulating cardiovascular remodeling[24-26]. Heterozygous KLF5-knockout mice showed reduced responses to cardiac injury, angiogenesis, hypertrophy and fibrosis[24,25]. In addition, KLF5 activity is regulated by other transcriptional regulators and nuclear receptors that are also involved in cardiovascular remodeling and injury response[24,25]. In tumor biology, KLF5 also has context-dependent proliferative or anti-proliferative activities in cancer cells and may function as either a tumor suppressor or an oncoprotein[27-29].
The functions of KLF5 are tightly controlled by post-translational modifications, including ubiquitination, SUMOylation, acetylation and phosphorylation[3-8,21,30,31]. For example, the SUMOylation of Lys151 and Lys202 regulates KLF5 nuclear localization[3]. Phosphorylation of KLF5 by PKC may enhance the transcriptional activities of KLF5 by promoting its interaction with CREB-binding protein[21]. In addition, KLF5 activity is also regulated by its acetylation status[4]. Moreover, KLF5 is a short-lived protein in cells and its protein level is tightly controlled by the ubiquitin-proteasome system[5-8,31,32]. Several E3 ubiquitin ligases, such as Smurf2, WWP1 and EFP, have been shown to degrade KLF5[7,31,32]. In 2010, Dr. Chen C’s group and our laboratory both reported that KLF5 is targeted for ubiquitination and degradation by the E3 ubiquitin ligase FBW7[6,8]. In the past three years, several studies from different groups have also provided evidence strongly supporting KLF5 as an essential FBW7 substrate under both physiological and pathological conditions[6-8,31-34].
Cellular protein levels are tightly controlled by protein degradation. The ubiquitin-proteasome system (UPS) is the major pathway for the degradation of approximately 90% of all proteins in cells[35-37]. The UPS acts by promoting protein ubiquitination and delivering the ubiquitinated proteins to the 26S proteasome for degradation[36]. The UPS is an enzymatic cascade containing three enzymes: enzyme-1 (E1), the ubiquitin-activating enzyme; E2, the ubiquitin carrier protein (ubiquitin-conjugating enzyme); and E3, the ubiquitin-protein ligase. E3 determines the specificity of protein degradation[35]. To date, more than 600 E3s have been identified in mammals and categorized into either the RING or HECT family of E3 ubiquitin ligases[38-40].
FBW7 (F-box and WD repeat domain-containing 7, also named CDC4, SEL10, or AGO) is the substrate recognition subunit of the E3 ubiquitin ligase complex SCFFBW7 (Skp1-Cullin-FBW7), which can target various proteins that are involved in cell proliferation for degradation[9]. Many substrates of FBW7 have been identified, including c-Myc, Cyclin E, Notch, TGIF, c-Jun, Mcl-1, p100 and so on (Table 1)[41-56]. There are three known isoforms of FBW7 with different subcellular localizations, including FBW7α, FBW7β and FBW7γ[9,57]. FBW7α is mainly localized to the nucleoplasm. FBW7β contains a transmembrane domain and is localized to the cytosol. FBW7γ is localized to the nucleolus via a nucleolar localization signal at its N terminus[9]. Each FBW7 isoform contains a F-box domain and WD40 repeats. The F-box domain contains approximately 40 amino acids that are involved in recruiting the SCF complex through direct interaction with SKP1. WD40 repeats are thought to form multiple contacts with various substrates[57-62].
| Substrate | Cdc4 phospho-degron | Phospho-site |
| CyclinE | LLTPPQSG | T380 S384 |
| Myc | LPTPPLSP | T58 S62 |
| JUN | GETPPLSP | T239 S243 |
| NOTCH1 | FLTPSPE | T2512 |
| TGIF | FNTPPPTP | T235 T239 |
| SRC3 | VHSPMASS | S505 S509 |
| mTOR | LLTPSIHL | T631 |
| MCL1 | DGSLPSTP | S159 T163 S121 |
| KLF5 | LNTPDLDM/PPSPPSSE/ NLTPPPSY | T244 S303 T324 |
| KLF2 | PDTPPLSPD/LLTPPSSP | T171 S175 T243 S247 |
| SREBP | TLTPPPSDAGSP | T426 S430 S434 |
| SV40 large T antigen | PPTPPPEP | T701 |
| MED13/MED13L | SSVTLTPPTS | T326 |
| NF-κB2 | LPSPPTSDSDSD | S707 S711 |
| C/EBPα | HPTPPPTP | T222 T226 |
| C/EBPσ | QPTPPQSP | T157 S161 |
| HIF1a | DQTPSPSDGSTRQSS | T497 S451 |
| AuroraA | LSYCHSK/NSSKPSN | S245 S387 |
| C-Myb | LMTPVSED | T572 S556 S528 |
| NRF1 | LFSPEVE | S350 |
| PGC1 | PLTPESPN/GLTPPTTP | T263 T295 |
FBW7 recognizes its substrates through a conserved phospho-epitope known as the Cdc4 phospho-degron (CPD), in which a central phospho-threonine/serine is embedded within hydrophobic residues in a I/L-I/L/P-pT-P-<K/R>4 (where K and R are unfavorable residues at positions 2 to 5) motif[9]. Most of the FBW7 substrates contain at least one conserved CPD, and the phosphorylation of the central Ser/Thr is usually mediated by the protein kinase GSK3β[61,63,64].
Numerous studies have demonstrated that FBW7 functions as a tumor suppressor in various cancers. Mutant FBW7 is frequently found in human tumors. For example, amino acid substitutions such as Q264R, H460R, and R465C have been found in breast cancer, cholangiocarcinoma and colon cancer, respectively[52,65-67].
KLF5 contains several potential CPDs[6]. Data from Dr. Chen’s group and our laboratory have indicated that all three isoforms of FBW7 can bind to KLF5 in vivo[6,8]. Mass spectrometry data have also shown that endogenous KLF5 can be co-purified with FBW7 in different cell types[46]. The interaction of KLF5 with FBW7 is dependent on the KLF5 CPD(s). Mutations within the KLF5 CPDs were shown to abolish the interaction. In addition, FBW7 binds to KLF5 via the WD40 repeats on FBW7. This interaction is also dependent on the phosphorylation of KLF5 CPDs by Glycogen synthase kinase 3 (GSK-3)β, and inhibition of GSK3β activity can reduce FBW7 binding to KLF5. GSK3β activity is regulated by various extracellular stimuli such as Wnt and growth factors[68,69], but it is still unclear whether the interaction between KLF5 and FBW7 is also regulated by extracellular signals.
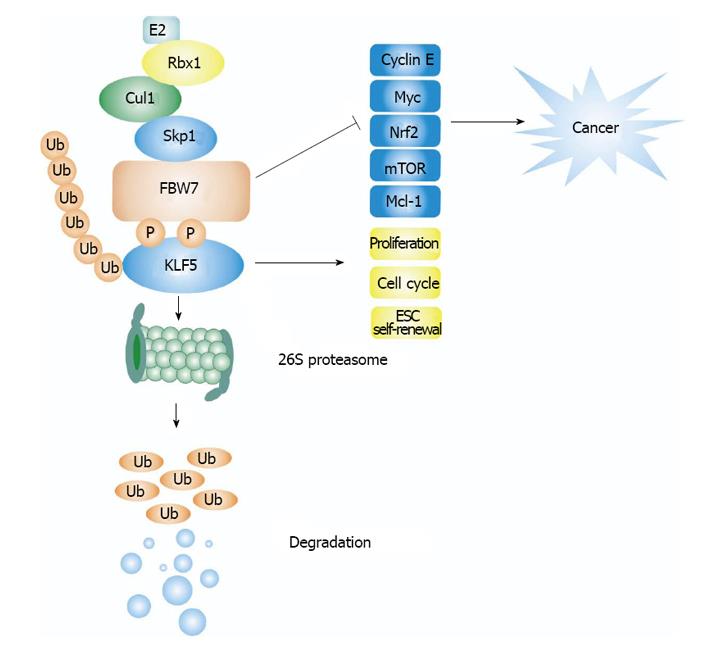
As a component of the SCF E3 ubiquitin ligase complex, co-expression of FBW7α or FBW7γ was shown to markedly promote the degradation of co-expressed KLF5, which could be blocked by the proteasome inhibitor MG132. In contrast, other F-box-containing proteins such as β-TrCP1, FBXW2, FBXW5 and FBXW8 had little effect on KLF5 stability. FBW7 with its F-box domain deleted or the WD40 domain of FBW7 alone failed to mediate KLF5 degradation, suggesting that FBW7-mediated KLF5 degradation requires the recruitment of other components of SCF E3 ligase. R338 residue in FBW7 is considered as a key residue in regulating the interaction of FBW7 with its substrates. Mutation of R338 to lysine blocks FBW7 mediated KLF5 degradation (Figure 2). Depletion of endogenous FBW7 significantly increased the amount of endogenous KLF5 protein without affecting the KLF5 mRNA level. KLF5 protein level was also upregulated in FBW7-deficient DLD1 cells and the half-life of endogenous KLF5 was dramatically extended in these cells compared with the WT DLD1 cells.
Moreover, FBW7 also promotes KLF5 ubiquitination in vitro and in vivo. The ubiquitination of KLF5 by FBW7 is dependent on the phosphorylation of KLF5 CPDs. Mutation of KLF5 CPDs dramatically blocked FBW7-induced KLF5 ubiquitination.
In addition to FBW7, WWP1, EFP and Smurf2 were also identified as E3 ligases that can target KLF5 for degradation[7,31,32]. Both WWP1 and Smurf2 belong to the HECT E3 ubiquitin ligase family[70,71]. Unlike FBW7, WWP1 and Smurf2 degrade KLF5 in a phosphorylation-independent manner. Interestingly, FBW7 and WWP1 appear to degrade KLF5 in a compensatory manner because knockdown of WWP1 was shown to cause an increase in FBW7 expression, and vice versa[8]. Degradation of KLF5 by multiple E3 ubiquitin ligases signifies the importance of the regulation of KLF5 protein stability under various physiological and pathological conditions[5-8,31-34].
FBW7 targets a substrate for degradation through the CPD consensus sites on the substrate[63]. KLF5 contains three potential CPDs: 242-LNTPDLDM, 301-PPSPPSSE and 322-NLTPPPSY (Table 1). Mutations of individual CPDs in mouse KLF5 were shown to have a minor effect on FBW7-mediated degradation. However, simultaneous mutations of two CPDs markedly blocked KLF5 interaction with FBW7 and KLF5 degradation. Mutations of all three CPDs completely abolished FBW7-induced KLF5 ubiquitination and degradation. Although KLF5 contains three CPDs, both Dr. Chen’s group and ours have found that phosphorylation of Ser303 in 301-PPSPPSSE is especially essential for FBW7-mediated degradation. In addition, Dr. Vincent W Yang’s group also found that P301 in KLF5 CPD is important for interaction between FBW7 and KLF5 and FBW7-mediated degradation of KLF5. P301S KLF5, a somatic mutation in KLF5 found in human colorectal cancer tissues, has a higher transcriptional activity than WT KLF5 and is resistant to FBW7α-mediated degradation, suggesting that P301S KLF5 mutant play an oncogenic role in colorectal cancer[72].
GSK-3 is a serine/threonine protein kinase[73] that phosphorylates the central serine/threonine residues in the CPDs of numerous FBW7 substrates[9], including KLF5. Co-expression of KLF5 with GSK3β was shown to promote KLF5 phosphorylation and KLF5 interaction with FBW7. Data from in vitro phosphorylation assays indicated that phosphorylation of wild-type KLF5 by GSK3β was much greater than that of a CPD-deficient KLF peptide, indicating that the KLF5 CPDs are phosphorylation targets of GSK3β. Inhibition of GSK3β by LiCl was shown to block FBW7-mediated KLF5 degradation. Conversely, KLF5 degradation was enhanced in the presence of the constitutively active GSK3β-S9A. Dr. Chen’s group reported similar results, and together these data indicate that GSK3β is required for FBW7-mediated degradation of KLF5.
Protein phosphorylation by GSK3β requires the phosphorylation of the priming phosphate group on a Ser/Thr residue that is located at the +4 position of a target residue[63]. For example, phosphorylation of c-Myc at T58 by GSK3β requires prior mitogen-activated protein kinase-dependent phosphorylation at serine S62[74-77]. Two of the KLF5 CPDs, 301-PPSPPSSE and 322-NLTPPPSY, contain a Ser at the +4 position. The protein kinase(s) that is involved in the phosphorylation of priming sites on KLF5 CPDs is still unknown.
We have previously shown that FBW7 negatively regulates the biological activity of KLF5[6]. An earlier study has also shown that KLF5 promotes the growth and proliferation of colorectal cancer cells[78]. Co-expression of FBW7 with KLF5 significantly inhibited the wild-type KLF5-mediated cell proliferation but had little effect on the proliferation of cells containing a CPD-mutant KLF5[6]. FBW7 can also inhibit the expression of KLF5 target genes, such as survivin, which regulates mitosis and caspase activity[79]. A high level of KLF5 has also been correlated with low survival in breast cancer patients[28]. Dr. Chen and his colleagues have determined the expression of FBW7 and KLF5 in multiple cancer cell lines, including HeLa, MCF10A, and 184B5 cells. Interestingly, they found that degradation of KLF5 by FBW7 is dependent on both the cell type and the FBW7 isoform[8]. For example, in 184B5 mammary gland cells, knockdown of FBW7α but not of the FBW7β and FBW7γ isoforms, upregulated the expression of KLF5 and its downstream target FGF-BP, which is a known promoter of breast cancer cell proliferation[8,80], suggesting that the different isoforms of FBW7 specifically regulate KLF5 stability and activity in breast cells.
Recently, several lines of evidence from mouse models indicate that KLF5 stability can be regulated by FBW7 in vivo[33,34,81]. As mentioned above, mutations of FBW7 occur frequently in multiple cancers, including those of the lung, colorectum, stomach, blood, pancreas, and endometrium. FBW7 R482Q is one of the loss-of-function mutants that have been identified in various cancers. A mouse model harboring the R482Q mutation was generated in Dr. Ian Tomlinson’s laboratory. Interestingly, the protein levels of KLF5 and TGIF1 were upregulated in the lungs of the heterozygous mutant mice, but the mRNA levels of these two genes remained the same between the mutant and the wild type mice[33,34]. Further investigation revealed that the levels of KLF5 and TGIF1 were also upregulated in normal intestine and adenomas of FBW7-deficient or FBW7-mutant mice. These data serve as strong in vivo evidences for KLF5 regulation by FBW7.
Regulation of KLF5 target gene expression by FBW7 has also been demonstrated in a mouse model[81]. Kumadaki et al[81] showed that in vivo knockdown of FBW7 significantly increased the hepatic expression of PPARγ2 as well as its targeted genes. More importantly, the degradation of KLF5 by FBW7 was associated with the inhibition of PPARγ2 expression. Thus, these findings suggested that degradation of KLF5 by FBW7 contributes to hepatic lipid metabolism.
In summary, FBW7 is an E3 ubiquitin ligase for KLF5. KLF5 contains functional CPDs that are phosphorylated by GSK3β, thus promoting the interaction between KLF5 and the WD40 domain of FBW7. This interaction subsequently leads to KLF5 ubiquitination and degradation by the ubiquitin-proteasome system. Mutation or deletion of FBW7 in cancer cells results in increased level of the KLF5 protein due to impaired degradation of KLF5, which in turn causes increased expression of KLF5 target genes, many of which can promote cell proliferation. Moreover, the KLF5 protein level is tightly controlled by FBW7 under normal physiological conditions, thus affecting many developmental and metabolic processes. In summary, the FBW7-KLF5 axis is important for both normal cellular activities, such as lipid metabolism, and cancer cell proliferation. This pathway may therefore serve as a novel target for cancer therapy
| 1. | McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010;90:1337-1381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 832] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 2. | Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66:2691-2706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 232] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 3. | Du JX, Bialkowska AB, McConnell BB, Yang VW. SUMOylation regulates nuclear localization of Krüppel-like factor 5. J Biol Chem. 2008;283:31991-32002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 4. | Matsumura T, Suzuki T, Aizawa K, Munemasa Y, Muto S, Horikoshi M, Nagai R. The deacetylase HDAC1 negatively regulates the cardiovascular transcription factor Krüppel-like factor 5 through direct interaction. J Biol Chem. 2005;280:12123-12129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 5. | Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, Simons JW, Dong JT. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319-3327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 115] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 6. | Liu N, Li H, Li S, Shen M, Xiao N, Chen Y, Wang Y, Wang W, Wang R, Wang Q. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285:18858-18867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 7. | Du JX, Hagos EG, Nandan MO, Bialkowska AB, Yu B, Yang VW. The E3 ubiquitin ligase SMAD ubiquitination regulatory factor 2 negatively regulates Krüppel-like factor 5 protein. J Biol Chem. 2011;286:40354-40364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70:4728-4738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 126] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 9. | Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 811] [Cited by in RCA: 886] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 10. | Dang DT, Pevsner J, Yang VW. The biology of the mammalian Krüppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103-1121. [PubMed] |
| 11. | Turner J, Crossley M. Mammalian Krüppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci. 1999;24:236-240. [PubMed] |
| 12. | Lania L, Majello B, De Luca P. Transcriptional regulation by the Sp family proteins. Int J Biochem Cell Biol. 1997;29:1313-1323. [PubMed] |
| 13. | Bieker JJ. Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355-34358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 497] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 14. | Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. [PubMed] |
| 15. | Black AR, Black JD, Azizkhan-Clifford J. Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 779] [Cited by in RCA: 856] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 16. | Sogawa K, Imataka H, Yamasaki Y, Kusume H, Abe H, Fujii-Kuriyama Y. cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucleic Acids Res. 1993;21:1527-1532. [PubMed] |
| 17. | Conkright MD, Wani MA, Anderson KP, Lingrel JB. A gene encoding an intestinal-enriched member of the Krüppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. 1999;27:1263-1270. [PubMed] |
| 18. | Shi H, Zhang Z, Wang X, Liu S, Teng CT. Isolation and characterization of a gene encoding human Kruppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res. 1999;27:4807-4815. [PubMed] |
| 19. | Ohnishi S, Ohnami S, Laub F, Aoki K, Suzuki K, Kanai Y, Haga K, Asaka M, Ramirez F, Yoshida T. Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308:251-256. [PubMed] |
| 20. | Chen C, Benjamin MS, Sun X, Otto KB, Guo P, Dong XY, Bao Y, Zhou Z, Cheng X, Simons JW. KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU-Pr1 human bladder cancer cell line. Int J Cancer. 2006;118:1346-1355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 126] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Zhang Z, Teng CT. Phosphorylation of Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function. Nucleic Acids Res. 2003;31:2196-2208. [PubMed] |
| 22. | Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 590] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 23. | Parisi S, Passaro F, Aloia L, Manabe I, Nagai R, Pastore L, Russo T. Klf5 is involved in self-renewal of mouse embryonic stem cells. J Cell Sci. 2008;121:2629-2634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 116] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 24. | Nagai R, Suzuki T, Aizawa K, Shindo T, Manabe I. Significance of the transcription factor KLF5 in cardiovascular remodeling. J Thromb Haemost. 2005;3:1569-1576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 80] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H. Krüppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8:856-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 323] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 26. | Santulli G, Basilicata MF, De Simone M, Del Giudice C, Anastasio A, Sorriento D, Saviano M, Del Gatto A, Trimarco B, Pedone C. Evaluation of the anti-angiogenic properties of the new selective αVβ3 integrin antagonist RGDechiHCit. J Transl Med. 2011;9:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | McConnell BB, Klapproth JM, Sasaki M, Nandan MO, Yang VW. Krüppel-like factor 5 mediates transmissible murine colonic hyperplasia caused by Citrobacter rodentium infection. Gastroenterology. 2008;134:1007-1016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 28. | Tong D, Czerwenka K, Heinze G, Ryffel M, Schuster E, Witt A, Leodolter S, Zeillinger R. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. 2006;12:2442-2448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Yagi N, Manabe I, Tottori T, Ishihara A, Ogata F, Kim JH, Nishimura S, Fujiu K, Oishi Y, Itaka K. A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo. Cancer Res. 2009;69:6531-6538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 30. | Miyamoto S, Suzuki T, Muto S, Aizawa K, Kimura A, Mizuno Y, Nagino T, Imai Y, Adachi N, Horikoshi M. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol Cell Biol. 2003;23:8528-8541. [PubMed] |
| 31. | Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553-41561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 120] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 32. | Zhao KW, Sikriwal D, Dong X, Guo P, Sun X, Dong JT. Oestrogen causes degradation of KLF5 by inducing the E3 ubiquitin ligase EFP in ER-positive breast cancer cells. Biochem J. 2011;437:323-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 33. | Davis H, Lewis A, Behrens A, Tomlinson I. Investigation of the atypical FBXW7 mutation spectrum in human tumours by conditional expression of a heterozygous propellor tip missense allele in the mouse intestines. Gut. 2014;63:792-799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 34. | Davis H, Lewis A, Spencer-Dene B, Tateossian H, Stamp G, Behrens A, Tomlinson I. FBXW7 mutations typically found in human cancers are distinct from null alleles and disrupt lung development. J Pathol. 2011;224:180-189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 35. | Hershko A. Ubiquitin: roles in protein modification and breakdown. Cell. 1983;34:11-12. [PubMed] |
| 37. | Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679-690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 688] [Cited by in RCA: 651] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 38. | Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1051] [Cited by in RCA: 1152] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 39. | Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1564] [Cited by in RCA: 1770] [Article Influence: 84.3] [Reference Citation Analysis (0)] |
| 40. | Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 819] [Cited by in RCA: 887] [Article Influence: 40.3] [Reference Citation Analysis (1)] |
| 41. | Bengoechea-Alonso MT, Ericsson J. A phosphorylation cascade controls the degradation of active SREBP1. J Biol Chem. 2009;284:5885-5895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 115] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 42. | Kwon YW, Kim IJ, Wu D, Lu J, Stock WA, Liu Y, Huang Y, Kang HC, DelRosario R, Jen KY. Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res. 2012;10:834-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 43. | Lee EC, Frolov A, Li R, Ayala G, Greenberg NM. Targeting Aurora kinases for the treatment of prostate cancer. Cancer Res. 2006;66:4996-5002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 141] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 44. | Yumimoto K, Matsumoto M, Onoyama I, Imaizumi K, Nakayama KI. F-box and WD repeat domain-containing-7 (Fbxw7) protein targets endoplasmic reticulum-anchored osteogenic and chondrogenic transcriptional factors for degradation. J Biol Chem. 2013;288:28488-28502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 45. | Kanei-Ishii C, Nomura T, Takagi T, Watanabe N, Nakayama KI, Ishii S. Fbxw7 acts as an E3 ubiquitin ligase that targets c-Myb for nemo-like kinase (NLK)-induced degradation. J Biol Chem. 2008;283:30540-30548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 46. | Davis MA, Larimore EA, Fissel BM, Swanger J, Taatjes DJ, Clurman BE. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 2013;27:151-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 102] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 47. | Tan M, Zhao Y, Kim SJ, Liu M, Jia L, Saunders TL, Zhu Y, Sun Y. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev Cell. 2011;21:1062-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 48. | Biswas M, Phan D, Watanabe M, Chan JY. The Fbw7 tumor suppressor regulates nuclear factor E2-related factor 1 transcription factor turnover through proteasome-mediated proteolysis. J Biol Chem. 2011;286:39282-39289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 49. | Fukushima H, Matsumoto A, Inuzuka H, Zhai B, Lau AW, Wan L, Gao D, Shaik S, Yuan M, Gygi SP. SCF(Fbw7) modulates the NFkB signaling pathway by targeting NFkB2 for ubiquitination and destruction. Cell Rep. 2012;1:434-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 50. | Busino L, Millman SE, Scotto L, Kyratsous CA, Basrur V, O’Connor O, Hoffmann A, Elenitoba-Johnson KS, Pagano M. Fbxw7α- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat Cell Biol. 2012;14:375-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 151] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 51. | Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 157] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 52. | Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006-9012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 395] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 53. | Flügel D, Görlach A, Kietzmann T. GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood. 2012;119:1292-1301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 167] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 54. | Kim DS, Zhang W, Millman SE, Hwang BJ, Kwon SJ, Clayberger C, Pagano M, Krensky AM. Fbw7γ-mediated degradation of KLF13 prevents RANTES expression in resting human but not murine T lymphocytes. Blood. 2012;120:1658-1667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 55. | Galli F, Rossi M, D’Alessandra Y, De Simone M, Lopardo T, Haupt Y, Alsheich-Bartok O, Anzi S, Shaulian E, Calabrò V. MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J Cell Sci. 2010;123:2423-2433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 56. | Welcker M, Clurman BE. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. J Biol Chem. 2005;280:7654-7658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 57. | Spruck CH, Strohmaier H, Sangfelt O, Müller HM, Hubalek M, Müller-Holzner E, Marth C, Widschwendter M, Reed SI. hCDC4 gene mutations in endometrial cancer. Cancer Res. 2002;62:4535-4539. [PubMed] |
| 58. | Matsumoto A, Onoyama I, Nakayama KI. Expression of mouse Fbxw7 isoforms is regulated in a cell cycle- or p53-dependent manner. Biochem Biophys Res Commun. 2006;350:114-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 59. | Kimura T, Gotoh M, Nakamura Y, Arakawa H. hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci. 2003;94:431-436. [PubMed] |
| 60. | Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263-274. [PubMed] |
| 61. | Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243-256. [PubMed] |
| 62. | Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 386] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 63. | Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 616] [Cited by in RCA: 619] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 64. | Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455-460. [PubMed] |
| 65. | Wei G, Wang Y, Zhang P, Lu J, Mao JH. Evaluating the prognostic significance of FBXW7 expression level in human breast cancer by a meta-analysis of transcriptional profiles. J Cancer Sci Ther. 2012;4:299-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 66. | Ibusuki M, Yamamoto Y, Shinriki S, Ando Y, Iwase H. Reduced expression of ubiquitin ligase FBXW7 mRNA is associated with poor prognosis in breast cancer patients. Cancer Sci. 2011;102:439-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 67. | Grim JE. Fbxw7 hotspot mutations and human colon cancer: mechanistic insights from new mouse models. Gut. 2014;63:707-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 68. | Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1175] [Cited by in RCA: 1258] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 69. | Jope RS, Yuskaitis CJ, Beurel E. Glycogen synthase kinase-3 (GSK3): inflammation, diseases, and therapeutics. Neurochem Res. 2007;32:577-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 579] [Cited by in RCA: 638] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 70. | Chen C, Zhou Z, Ross JS, Zhou W, Dong JT. The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer. 2007;121:80-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 108] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 71. | David D, Nair SA, Pillai MR. Smurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppression. Biochim Biophys Acta. 2013;1835:119-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 72. | Bialkowska AB, Liu Y, Nandan MO, Yang VW. A colon cancer-derived mutant of Krüppel-like factor 5 (KLF5) is resistant to degradation by glycogen synthase kinase 3β (GSK3β) and the E3 ubiquitin ligase F-box and WD repeat domain-containing 7α (FBW7α). J Biol Chem. 2014;289:5997-6005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 73. | Cohen P, Goedert M. GSK3 inhibitors: development and therapeutic potential. Nat Rev Drug Discov. 2004;3:479-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 638] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 74. | Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501-2514. [PubMed] |
| 75. | Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 626] [Article Influence: 28.5] [Reference Citation Analysis (6)] |
| 76. | Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116-2125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 551] [Cited by in RCA: 678] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 77. | Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085-9090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 711] [Cited by in RCA: 779] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 78. | Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216-1221. [PubMed] |
| 79. | Cheung CH, Huang CC, Tsai FY, Lee JY, Cheng SM, Chang YC, Huang YC, Chen SH, Chang JY. Survivin - biology and potential as a therapeutic target in oncology. Onco Targets Ther. 2013;6:1453-1462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 80. | Zheng HQ, Zhou Z, Huang J, Chaudhury L, Dong JT, Chen C. Krüppel-like factor 5 promotes breast cell proliferation partially through upregulating the transcription of fibroblast growth factor binding protein 1. Oncogene. 2009;28:3702-3713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 81. | Kumadaki S, Karasawa T, Matsuzaka T, Ema M, Nakagawa Y, Nakakuki M, Saito R, Yahagi N, Iwasaki H, Sone H. Inhibition of ubiquitin ligase F-box and WD repeat domain-containing 7α (Fbw7α) causes hepatosteatosis through Krüppel-like factor 5 (KLF5)/peroxisome proliferator-activated receptor γ2 (PPARγ2) pathway but not SREBP-1c protein in mice. J Biol Chem. 2011;286:40835-40846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
P- Reviewers: Choi CY, Santulli G, Tomita Y S- Editor: Gou SX L- Editor: A E- Editor: Lu YJ