Published online Feb 21, 2019. doi: 10.4331/wjbc.v10.i2.28
Peer-review started: November 29, 2018
First decision: December 24, 2018
Revised: January 2, 2019
Accepted: January 28, 2019
Article in press: January 28, 2019
Published online: February 21, 2019
Processing time: 84 Days and 20.9 Hours
Fasciculation and elongation zeta/zygin (FEZ) proteins are a family of hub proteins and share many characteristics like high connectivity in interaction networks, they are involved in several cellular processes, evolve slowly and in general have intrinsically disordered regions. In 1985, unc-76 gene was firstly described and involved in axonal growth in C. elegans, and in 1997 Bloom and Horvitz enrolled also the human homologues genes, FEZ1 and FEZ2, in this process. While nematodes possess one gene (unc-76), mammalians have one more copy (FEZ1 and FEZ2). Several animal models have been used to study FEZ family functions like: C. elegans, D. melanogaster, R. novergicus and human cells. Complementation assays were performed and demonstrated the function conservation between paralogues. Human FEZ1 protein is more studied followed by UNC-76 and FEZ2 proteins, respectively. While FEZ1 and UNC-76 shared interaction partners, FEZ2 evolved and increased the number of protein-protein interactions (PPI) with cytoplasmatic partners. FEZ proteins are implicated in intracellular transport, acting as bivalent cargo transport adaptors in kinesin-mediated movement. Especially in light of this cellular function, this family of proteins has been involved in several processes like neuronal development, neurological disorders, viral infection and autophagy. However, nuclear functions of FEZ proteins have been explored as well, due to high content of PPI with nuclear proteins, correlating FEZ1 expression to Sox2 and Hoxb4 gene regulation and retinoic acid signaling. These recent findings open new avenue to study FEZ proteins functions and its involvement in already described processes. This review intends to reunite aspects of evolution, structure, interaction partners and function of FEZ proteins and correlate them to physiological and pathological processes.
Core tip: Fasciculation and elongation zeta/zygin (FEZ) proteins are intrinsically disordered and hub proteins involved in many cellular functions, acting as a bivalent adaptor of kinesin-based movement. These proteins are associated to several processes like neuronal development, neurological disorders, viral infection and autophagy. However, novel nuclear functions are being described, shedding more light to their role. This review intends to reunite aspects of evolution, structure, interaction partners and function of FEZ proteins and correlate them to physiological and pathological processes.
- Citation: Teixeira MB, Alborghetti MR, Kobarg J. Fasciculation and elongation zeta proteins 1 and 2: From structural flexibility to functional diversity. World J Biol Chem 2019; 10(2): 28-43
- URL: https://www.wjgnet.com/1949-8454/full/v10/i2/28.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v10.i2.28
The fasciculation and elongation zeta/zygin (FEZ) protein family was described by Bloom and Horvitz[1] in 1997 as two human homologs of UNC-76 protein (Table 1). In attempt to characterize the unc-76 gene, which in Caenorhabditis elegans mutants caused locomotory defects (uncoordinated), they found that these mutants presented axonal abnormalities: axons in fascicles did not reach their full lengths, and also failed to bundle tightly together. In addition, human FEZ1 gene (protein code Q99689) was capable to partially restore unc-76 mutant locomotion defects and axonal fasciculation, thus suggesting that FEZ family members share conserved evolutionary function and structure from C. elegans to Homo sapiens[1].
| UNC-76 identity (%) | UNC-76 similarity (%) | FEZ1 identity (%) | FEZ1 similarity (%) | |
| FEZ1 | 35 | 46 | - | - |
| FEZ2 | 34 | 45 | 49 | 56 |
The worm has one copy of unc-76 gene, while humans have two copies, FEZ1 and FEZ2 (protein code Q9UHY8). It has been later proposed that unc-76 gene duplication occurred after divergence in the amphioxus branch, concomitant with chordates origin[2]. Synteny analysis evidences two rounds of genomic duplication in the chordate branch, after cephalochordate divergence but before the division of teleost and tetrapod[3]. Probably, the unc-76 gene duplication has occurred during these rounds of genomic duplication.
Bloom and Horvitz[1] in 1997 also gave some insights into FEZ1/UNC-76 structure, function and expression pattern, which during more than 20 years of research were - and still are - the main subjects of study from different groups around the world[1]. Further in this paper we will discuss these topics in details.
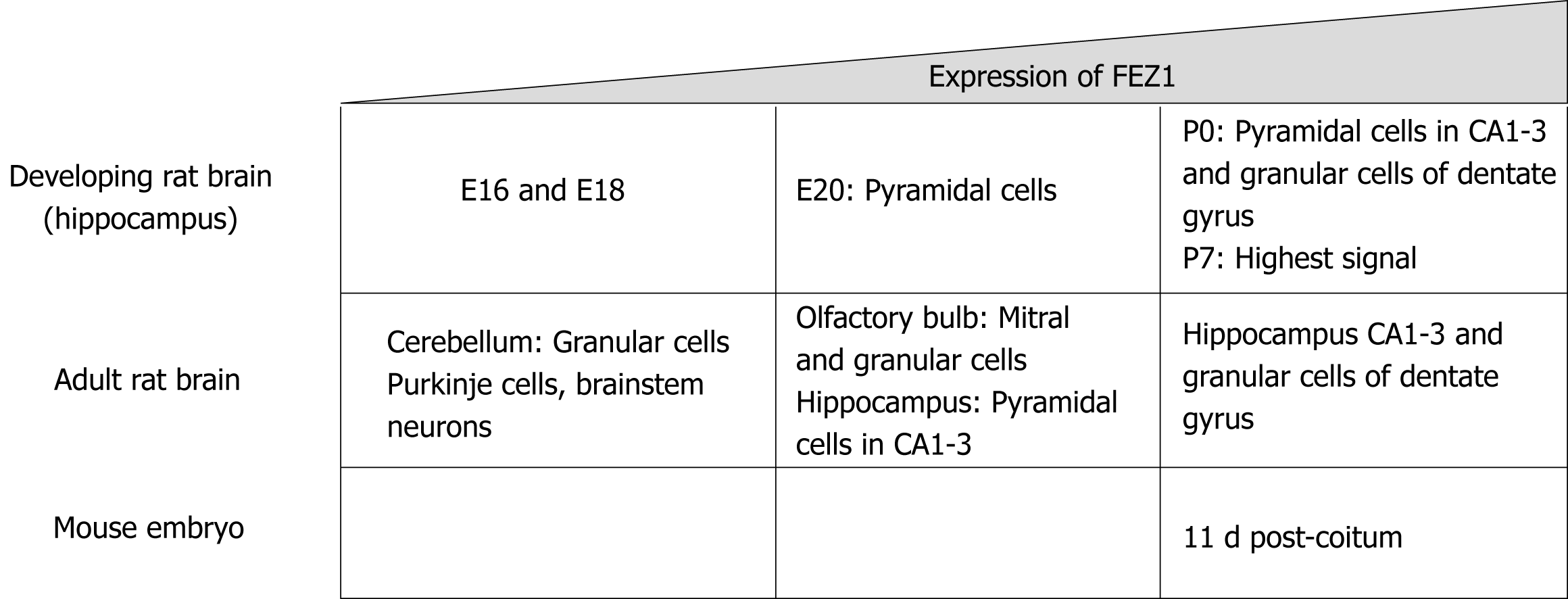
As previously stated, Bloom and Horvitz[1] in 1997 briefly reported the expression patterns regarding FEZ1 and FEZ2, with the former being present in the brain while the latter also in non-neuronal tissues. Later, Honda et al[4] in 2004 characterized the expression of FEZ1 in the developing rat brain by in situ hybridization. It was shown that FEZ1 mRNA in adult rat brain was more expressed in olfactory bulb and cortical and hippocampal neurons, while the signal in cerebellum was weak.
Regarding the expression levels during development in rat, FEZ1 mRNA expression was low in the hippocampus by E16 and E18 prenatal development stages, by E20 there was a signal in pyramidal cells, and by P0 there was an intense signal in both pyramidal cells of the CA1-3 regions and granule cells of the dentate gyrus. The highest signal of FEZ1 mRNA was detected at P7 and in adult rats the expression decreased[4].
Another study compared the mRNA expression levels of FEZ1 and FEZ2 in rat tissue and mouse embryos. FEZ1 mRNA was observed almost exclusively in the brain, while FEZ2 mRNA was ubiquitously present in all tissues, although weaker when compared to FEZ1. In mouse developing embryos, FEZ1 mRNA was greatly increased around 11 dpc (days post-coitum) and gradually faded as development continued. FEZ2 mRNA, otherwise, showed to be constantly expressed from 7 to 17 dpc[5]. Figure 1 presents a schematic view of FEZ1 expression.
Northern blot analysis with RNA from adult human tissues showed weak presence of FEZ1 RNA in prostate, testis, ovary, small intestine, colon, liver, especially when compared with very high expression of FEZ1 RNA in the brain[6].
Moreover, a gene array analysis of rat type-1 astrocytes (T1As) and T2As has also shown the expression of FEZ1 mRNA. At both mRNA and protein levels, T2As expressed more FEZ1 than T1As. Immunofluorescent staining with specific antibody showed the presence of FEZ1 in the cytoplasm of both astrocytes and also neurons[7]. More recently, the same group reported a new study using a rat model for Parkinson’s disease (PD) by injecting 6-Hydroxydopamine Hydrobromide in the medial forebrain bundle. The results showed an increase of FEZ1 mRNA and protein in both striatum and substantia nigra of PD rats, peaking at 2 and 3 wk respectively after injury, and then decreasing to control levels for striatum and also decreasing for substantia nigra but to levels that were still higher when compared to control rats. In addition, after performing immunostaining of brain tissue after 2 wk of injection, the authors observed a change of FEZ1 expression from dopamine neurons in control group to a significantly increase of its expression in astrocytes from PD rats, suggesting an association between FEZ1 expression and astrocyte activation after injury[8].
More recently, it has been reported that FEZ1 is expressed in oligodendroglia (OL) lineage cells originated from mouse, rat, and human, through immunofluorescence. In addition, the authors also found that protein and mRNA levels of FEZ1 were increased by day 4 of differentiation process in primary cultured oligodendroglia progenitor cells (OPCs) in comparison to undifferentiated proliferating OPCs. Moreover, upregulation of FEZ1 protein and mRNA was detected in optic nerves during intense myelinogenesis between postnatal days 14 and 30[9].
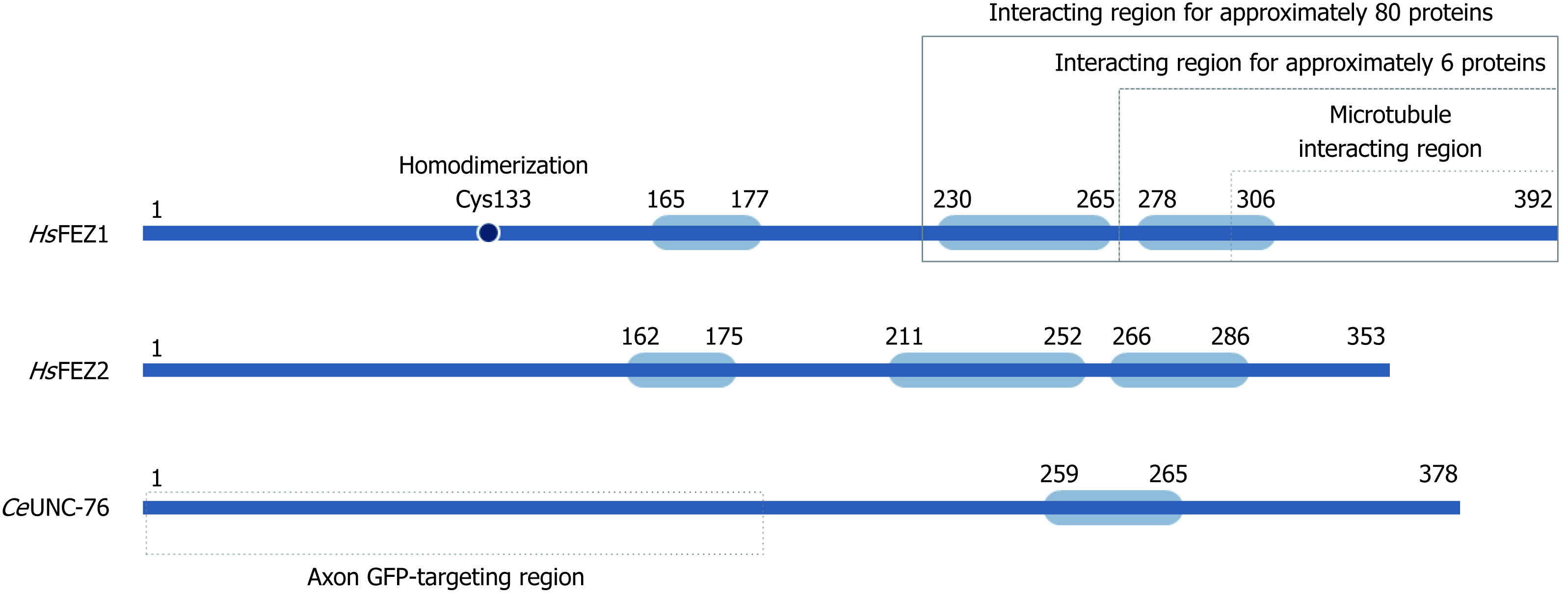
Human FEZ1 possess 392 amino acids and through different software predictions it was shown that its whole sequence, especially N-terminal and central regions, exhibited disordered structure. The C-terminal region was predicted to possess higher probability of structure. Indeed after performing circular dichroism spectro-polarimetry with purified protein, the authors confirmed the prediction of a largely disordered structure for FEZ1 protein and also suggested that it could gain structure upon binding to interacting partners[10].
It was also in silico predicted for FEZ1 to present one region with high probability to form coiled-coils in the regions between amino acid residues 230-265 (approximately 96% probability) and 278-306 (63%) (Figure 2). At the N-terminal three regions with low probability were also predicted[10]. Similar structure organization is found at homologues FEZ2 (human) and UNC-76 (C. elegans) (Figure 2).
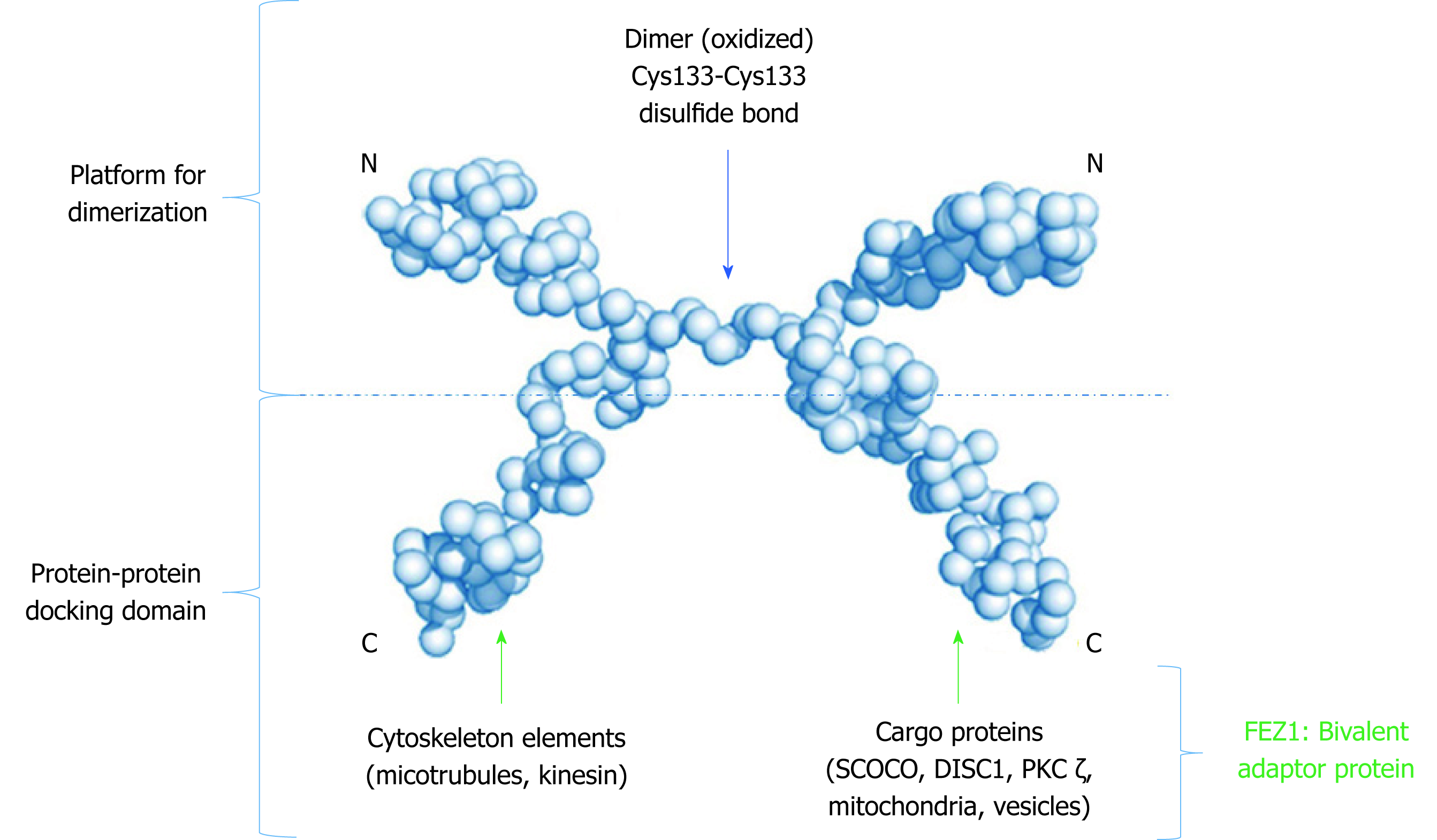
Due to its unstructured characteristic, small angle X-ray scattering (SAXS) experiments were performed to determine low 3D resolution conformational model of FEZ1. Besides showing that FEZ1 has a long, extended and flexible conformation, the dimeric state of the protein in solution was also demonstrated. The authors compared the theoretical value of molecular mass for full length FEZ1 (48.6 kDa) with the one of the dimer (approximately 95 kDa), confirming that the latter was twice the former[10]. It is also important to highlight, especially for didactic purpose that due to the high content of charged amino acids and intrinsically disordered structure, FEZ1 shows an anomalous mobility in SDS-PAGE.
A second publication about FEZ1 dimerization deepened our understanding on this matter. After also performing SAXS experiments, the authors confirmed that the N-terminal region (protein fragment 92-194 amino acids, contain coiled-coil motif) was able to dimerize in solution without a disulfide bond reducing agent, but become a monomer after its addition. Moreover, through mass spectroscopy in full length FEZ1, it was shown that the dimeric peptide was formed by a Cys133–Cys133 disulfide bond. The authors also confirmed the presence of endogenous FEZ1 as a dimer in HEK293 cell lysate. Regarding FEZ2 (N-terminal fragment 106-189 amino acids, with coiled-coil), due to low concentration expression, the dimer state was only analyzed by SDS-PAGE followed by Western blot. Two bands were shown: monomeric band of 20 kDa upon reduction and dimer band of 40 kDa under nonreducing condition[11].
Finally, the structure model proposed (Figure 3) by all these authors determines that FEZ1 forms a dimer at the N-terminal region through a disulfide bond and that the outwards pointing C-terminal regions from each monomer can freely interact with the many partner proteins of FEZ1 interactome (discussed below). Hence, the N-terminal serves as a platform for dimerization while the C-terminal works as a protein-protein docking domain.
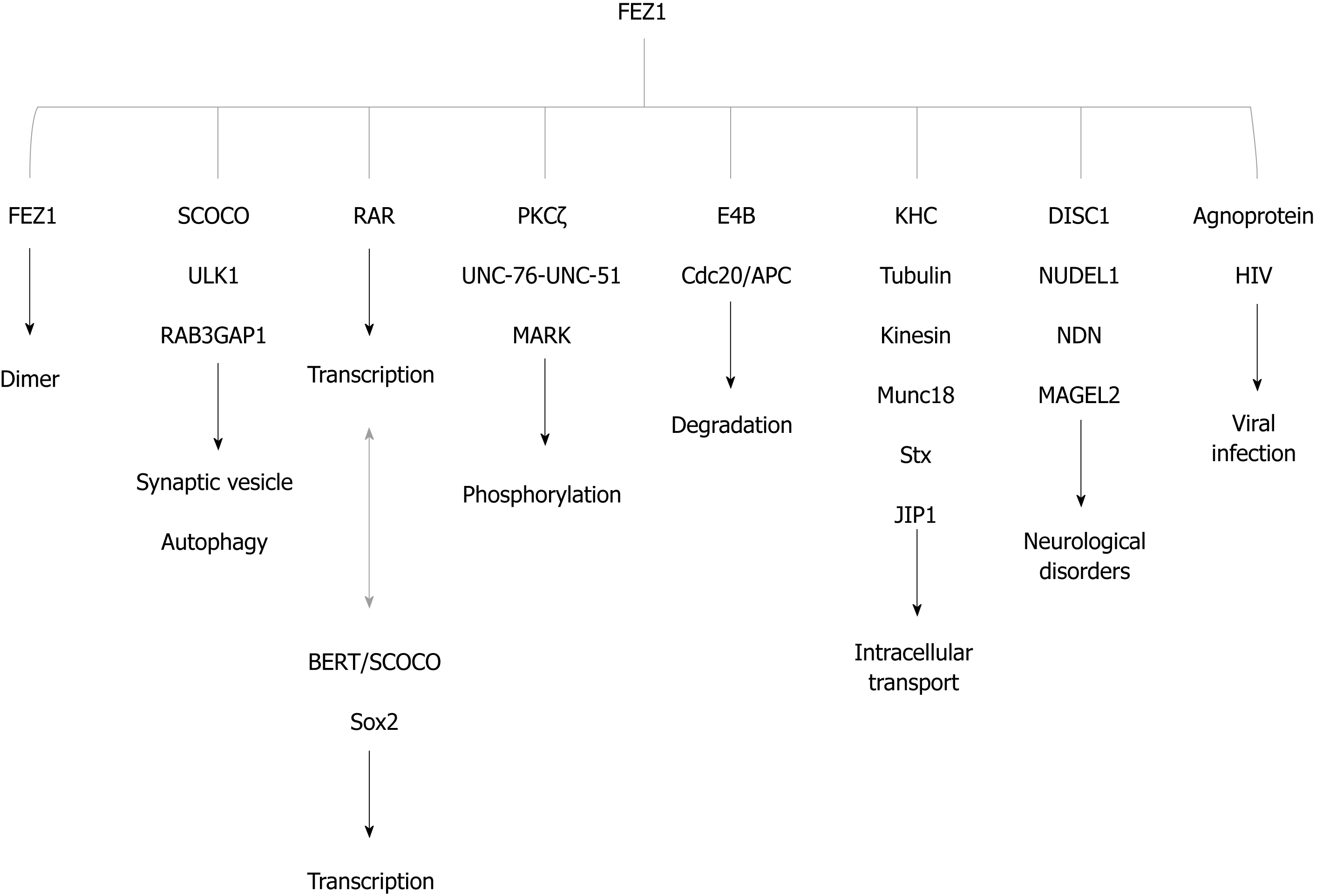
FEZ1 and FEZ2 are considered hub proteins, meaning that they interact with a great number of other proteins, which possess different functions (Figure 2). Makino et al[12] suggested that the evolution rate of a protein with many interaction partners is slower than those with few of them. After gene duplication, three functional pathways are possible. First, one copy can be silenced while the other copy remains with the original function. Second, while one copy remains with original function, the other copy can acquire new functions by accumulation of mutations that are positively selected. Third, both gene copies gain additional new functions, but the original function is cooperatively retained[12]. FEZ interaction partners indicate that the second option is currently happening with Fez genes.
Two independent yeast two-hybrid studies were performed to identify interaction partners (see details in: Assmann et al[13], 2006; Alborghetti et al[2], 2011). Here we summarized the main functions and interactors in Figure 4.
Alborghetti et al[2] in 2011 demonstrated that UNC-76 interacted with practically all FEZ1 interaction partners. FEZ2 was also able to interact with all FEZ1 partners and acquired new exclusive partners. The authors found that 83.3% of nuclear proteins interacting with FEZ2 also interacted with FEZ1. Proteins involved with translation seemed to be the novelty specific to FEZ2, and 56% of cytoplasmic proteins found interacting with FEZ2 were specific and did not interact with FEZ1[2].
This interaction pattern and the previously proposed history of unc-76 gene duplication are in accordance to phenotypes displayed in mutation, knockout and complementation studies in C. elegans, Drosophila melanogaster and Mus musculus. Defects in axonal development were observed in gene mutations in invertebrates and human FEZ1 was able to complement them in C. elegans. However, FEZ1 knockout mice did not show any impairment in axonal development, although these animals were more responsive to methamphetamine[1,14-16]. Since FEZ2 is able to interact with all FEZ1 interaction partners, it is possible, though still speculative, that FEZ2 can complement FEZ1 function in its absence.
The first interaction partner reported for FEZ1 was the protein kinase C zeta (PKCζ), which is an atypical isoform of the PKC family. Kuroda et al[17] in 1999 performed a yeast two-hybrid screening using a rat brain cDNA library and found that the regulatory domain of rat PKC zeta interacted with FEZ1, most likely in its C-terminal. The interaction was confirmed by immunoprecipitation in COS-7 cells overexpressing both FEZ1 and PKC zeta, and the authors found that the complex could be phosphorylated after performing a phosphorylation assay. One interesting aspect discovered was that transfected FEZ1 seemed to translocate from the plasma membrane and cytoplasm periphery to uniform cytoplasm localization when there was a change in overexpressing PKC zeta to a constitutively active form of PKC (caPKCζ). Moreover, it was also shown that PC12 cells’ rate of differentiation into neurons was higher (approximately 48%) in the presence of transfected FEZ1 and caPKCζ than when only caPKCζ was transfected (approximately 18%)[17]. A subsequent study showed that FEZ2 and PKCζ were immunoprecipitated and that PC12 could differentiate more in the presence of both FEZ2 and caPKCζ [5].
Still regarding the matter of phosphorylation, it was later reported that UNC-76 interacted and was phosphorylated by UNC-51, a Ser/Thr kinase, in Droshophila. The authors performed experiments in vitro, in HEK293T cells and in extracts from wild-type larvae in order to address the phosphorylation status. Mass spectrometry analysis revealed that UNC-76 Ser143 was a phosphorylation site. In addition, phosphorylated UNC-76 was detected in wild-type and absent in the unc-51 fly mutant. Also, a mutated version of phospho-UNC76 (S143A) could not rescue the defects present in unc-76 mutants, in opposition to the rescued effect caused by UNC-76 wild-type and a phosphomimetic UNC-76. Those defects were related to axonal transport and synaptic vesicles and kinesin heavy chain (KHC) aggregation. Moreover, co-expression in HEK293T cells followed by immunoprecipitation and FRET analysis in COS-7 cells, showed the interaction between UNC-76 and Synaptotagmin-1, a protein of synaptic vesicles, and that this interaction was dependent of UNC-76 phosphorylation by UNC-51. Drosophila unc-51 mutants produced a phenotype where axonal cargoes transport was affected, especially the one of synaptic vesicles and partially the mitochondria[18].
Using liquid chromatography coupled with tandem mass spectrometry, another work identified four FEZ1 phospho-serine sites (S58, S134, S301, S316) from HEK293 cells lysates expressing FEZ1-GFP[19]. The phosphorylation of the S58 site of FEZ1 by microtubule-affinity regulating kinases (MARKs) was confirmed in vitro and in HEK293 cells. Specific antibody raised against the residue S58 tested in HeLa cells confirmed that, in the presence of MARK, phospho-FEZ1 was detected[20].
Regarding the degradation of FEZ1, some studies shed light on how this could happen. Starting with a yeast two-hybrid assay using a human brain cDNA library, it was showed that FEZ1 interacted with the U-box-type ubiquitin ligase E4B. The interaction was confirmed in HEK293T and PC12 cells. The authors reported that the interaction was enhanced in the presence of caPKCζ. In addition, they found that co-expression of E4B with FEZ1 and caPKCζ increased the proportion of PC12 cells differentiated into neurons. Although there was some polyubiquitylation of FEZ1 by E4B, the authors could not directly affirm that this modification causes FEZ1 degradation by the proteasome[21]. Nonetheless, it was shown that the presence of the proteasome inhibitor MG132 prevented the degradation of FEZ1[22,23], indicating that FEZ1 is likely to be degraded by the 26S proteasome.
A more recent study showed that FEZ1 interacted with cell-division cycle 20/anaphase-promoting complex (Cdc20/APC), which is an ubiquitin ligase (E3) and modulates dendrite development. Experiments carried in vitro, in HEK293T cells and in hippocampal lysates confirmed the interaction. The authors reported that over-expression of Cdc20 increased the polyubiquitination of FEZ1 and that the suppression of Cdc20 increased the levels of FEZ1, suggesting that the degradation of FEZ1 is regulated by Cdc20 in dendrite development. In addition, the authors also reported that the Cdc20/APC complex is regulated by Hdac11[23]. A subsequent new study with Hdac11 knockout mice showed decrease of Fez1 in the hippocampus of adult mice[24].
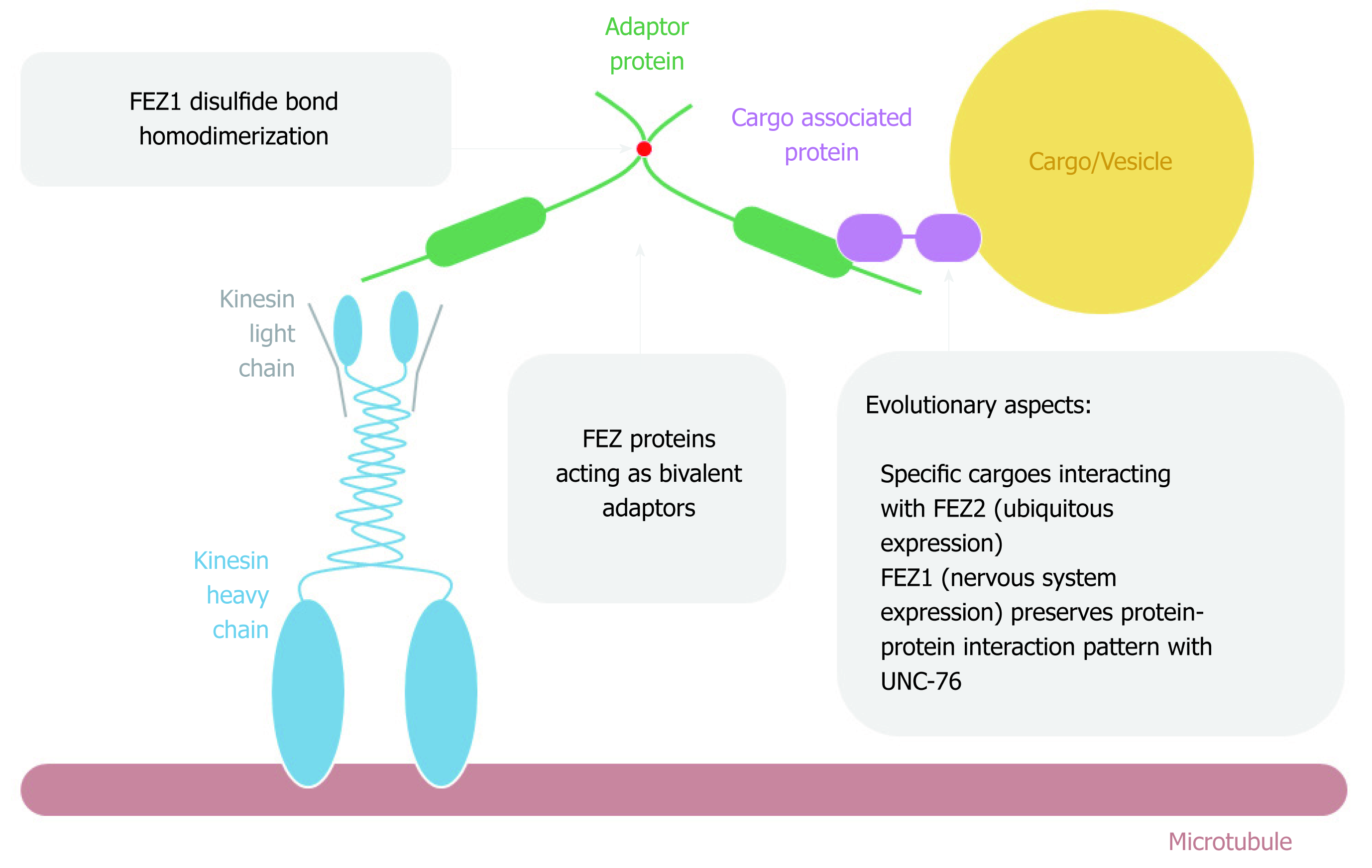
It was first reported by Gindhart et al[14] in 2003 that UNC-76 was found as an interaction partner of KHC tail domain after performing a two-hybrid assay using a Drosophila embryonic cDNA library. The interaction was further confirmed in vitro. A general scheme of FEZ proteins acting on cytoplasmatic intracellular transport is depicted on Figure 5. The authors generated unc-76 fly mutants and performed detailed characterization. They found that UNC-76 was essential for Drosophila development and the phenotypes were similar to the mutants for kinesin. The function of UNC-76 as a membrane cargo transporter in axons was not observed since there was no co-localization with axon clogs, which are related to vesicle transport[14].
Nonetheless, in line with this first study, subsequent work provided more evidence about FEZ1 as an important component of the kinesin transport pathway. In vitro experiments showed the interaction between the C-terminal of FEZ1-tubulin and FEZ1-kinesin, and the complex FEZ1-kinesin (KIF5)-β tubulin was immuno-precipitated from PC12 cells differentiated into neurons. The authors also showed the co-localization of endogenous FEZ1 and mitochondria in PC12 cells, both before and after differentiation into neurons upon treatment with nerve growth factor (NGF)[25]. They also confirmed another work[26] showing that mitochondria anterograde movement was reduced when FEZ1 was repressed by interference RNA, both in hippocampal neurons isolated from rat embryonic brain and PC12 cells treated with NGF[25]. Therefore, it was shown that FEZ1 is important in mitochondrial anterograde transport, morphology and neuronal polarity and this transport along microtubules was mediated by FEZ1-Kinesin interaction. More recently, it has been reported that UNC-76 regulated mitochondrial density in anterior C. elegans touch receptor neuron[27].
Another study identified a transport complex involving Kinesin-1 (KIF5C), FEZ1, Munc18 and Syntaxin 1a (Stx). Immunoprecipitation in HEK293 and also neurons from postnuclear supernatants from rat brain confirmed the function of FEZ1 as an adaptor that connected Munc18 and Stx (cargoes) to the kinesin-1 (motor). Phosphatase treatment ceased the interaction of FEZ1 with Munc18 and KIF5C. Stx and Munc18 colocalized with FEZ1 in growth cones of primary hippocampal neurons. C. elegans mutants for UNC-76 and UNC-116 (kinesin) presented aggregation of UNC-64 (Stx) in cells bodies and axons[19].
Using label-free quantitative mass spectrometry in immunoisolated FEZ1 and Kinesin-1 samples of the cytosolic fraction containing vesicles from the rat brain, it was found that over one thousand proteins were enriched in the samples. More surprisingly, more than 90% of these proteins were common for both FEZ1 and Kinesin-1. The authors also generated transgenic C. elegans and found that the disruption of Par-1 (the only MARKs in worms) altered the phenotype of FEZ1 and Stx1 with the appearance of aggregates in axons and cells bodies, probably due to impairment of axonal transport[20].
In addition, another component of the complex was found to be the protein c-Jun N-terminal kinase-interacting protein 1 (JIP1). After confirming the interaction between kinesin-1 (kinesin heavy and light chain) with FEZ1 and JIP1 in COS cells by co-immunoprecipitation (co-IP), the authors performed a microtubule binding assay showing that the presence of both FEZ1 and JIP1 was necessary for the activation of kinesin-1 and the microtubule motility. The presence of only one of the proteins could not activate kinesin-1 and, in the absence of kinesin-1, there was no interaction between FEZ1 and JIP1[28].
One of the genes related to schizophrenia encodes the protein Disrupted-In-Schizophrenia 1 (DISC1) and it was identified to interact with FEZ1 through a two-hybrid assay. More specifically, the C-terminal of both proteins was required for this interaction. In addition, the interaction was further confirmed by experiments in mammalian cells. The authors also demonstrated that during neuron differentiation in PC12 cells, the interaction FEZ1-DISC1 was crucial for neurite outgrowth[29]. Another study showed that the knockdown of FEZ1 in cultured adult mouse neural progenitors derived from hippocampus caused increased soma size of neurons and accelerated dendritic development, being both phenotypes also present in DISC1 knockdown neurons. Moreover, double knockdown experiment for both of these proteins confirmed an enhancement of the dendritic growth phenotype when compared to FEZ1 knockdown alone, suggesting a synergistic functional role of DISC1-FEZ1 interaction in regulating dendritic development[30].
In accordance, the work from Honda et al[4] in 2004 has previously reported similarities regarding the expression patterns of DISC1 and FEZ1 in rat brain during prenatal stages and at birth, suggesting a role for this interaction during the nervous system development in mammals. Moreover, it is noteworthy that the region containing the translocation breakpoint responsible for disrupting DISC1 in schizophrenia was critical for the interaction with FEZ1[29]. These findings motivated some interesting reports where genotyping of different patient populations was performed in order to associate Fez1 gene with schizophrenia. The study with a Japanese population showed this association for a specific group of patients carrying rare homozygosis[31]. A more robust study was further performed in the Japanese population and the former could not be replicated[32]. In accordance with the latter, a study conducted in Caucasian and African American populations could not associate any of the nine single nucleotide polymorphisms tested for Fez1 with schizophrenia[33].
Nevertheless, studies have continued to be performed in attempt to correlate FEZ1 with schizophrenia, especially in association with other genes. One work conducted with postmortem hippocampal and dorsolateral prefrontal cortex from normal controls and patients with schizophrenia revealed a significant decrease in FEZ1 mRNA expression in the test group[34]. More recently, another study has also shown the same down-regulation of FEZ1 mRNA levels in peripheral blood samples from patients with schizophrenia when compared to healthy volunteers[35]. In addition, FEZ1 mRNA showed to be a significant diagnosis predictor. The mRNA from nuclear distribution element-like (NUDEL) was also significantly decreased in hippocampus and proved to be useful as a diagnosis predictor[34]. Interestingly, it was later described a significant positive correlation between FEZ1 and NUDEL regarding their expression patterns in RNA samples obtained from postmortem human frontal cortex[36]. At protein level, co-IP experiments with adult mouse brain tissue confirmed the interaction between FEZ1/NUDEL1 and DISC-1/NUDEL1. Functionally, double knockdown of FEZ1 and NUDEL1 showed additive phenotype described for each individual protein knockdown, demonstrating different roles in regulation of neurogenesis in adult mouse brain[30].
Another aspect of FEZ’s association with neurological diseases is its interaction with proteins necdin (NDN) and MAGEL2, both inactivated in Prader-Willi syndrome and predicted to relate with hypothalamic dysfunction. The authors reported the interaction of FEZ2 with NDN by yeast two-hybrid Ras rescue system and also the interaction of both FEZ1 and FEZ2 with NDN by co-IP in HEK293 cells transiently transfected. There was also interaction of FEZ1 with MAGEL2 when performing co-IP. Immunofluorescence performed in HEK293 cells overexpressing NDN and FEZ1 showed co-localization of these proteins in juxtanuclear bodies as well as the presence of FEZ1 surrounding and partially overlapping the gamma-tubulin at the centrosomes. Co-IP of FEZ1, NDN and gamma-tubulin showed that these proteins were all present in the immuno complex. Together with other results, the authors proposed that the NDN-FEZ1 interaction in centrosomal structure could cause cytoskeletal rearrangements that interfere in axonal outgrowth[22].
The JC virus (JCV) agnoprotein is related to viral growth and JCV infection. Using a human brain cDNA library and the full-length agnoprotein, Suzuki et al[37] in 2005 performed a two-hybrid yeast assay and found FEZ1 as a binding partner of agnoprotein. The FEZ1 region retrieved contained the coiled-coil domain. Deletion mutants of FEZ1 and purified protein precipitation assays further confirmed the direct interaction of agnoprotein and the coiled-coil at C-terminal of FEZ1. Moreover, a stable cell line expressing JCV agnoprotein derived from HEK293 cells was transfected with FEZ1 and the interaction was also confirmed by immuno-precipitation. Confocal microscopy showed agnoprotein in the perinuclear region and also extended to the cytoplasm, while FEZ1 was seen throughout the cytoplasm and colocalized with agnoprotein only in the perinuclear region[37].
Functionally, it was found that agnoprotein significantly inhibited neurite extension in transfected PC12 cells stably expressing FEZ-GFP with DsRed-agnoprotein. Additionally, a human glial cells (SVG-A) stably expressing FEZ1 was inoculated with JCV and the expression of agnoprotein and VP1 (protein from the viral capsid) was reduced in the cells overexpressing FEZ1 when compared to control. Conversely, the same proteins showed increased expression in cells transfected with FEZ1 siRNA. Thus, FEZ1 suppressed the expression of JCV proteins in SVG-A cells. It was also shown by immunocytofluorescence in the same cell line that overexpression of FEZ1 suppressed the propagation of VP1-positive cells after 7 d of infection and that this viral protein was restricted to the nucleus in the presence of FEZ1, suggesting a role as an inhibitor of nuclear to cytoplasm translocation[37].
Interestingly both FEZ1 and agnoprotein were also shown to interact with microtubules through microtubule cosedimentation assays. The binding of FEZ1 and microtubules occurred at the C-terminal of FEZ1 (residues 297-392), which does not contain the coiled-coil region, meaning that binding regions of agnoprotein and microtubules with FEZ1 were different from each other. More importantly it was reported that agnoprotein and FEZ1 seemed to compete for microtubule: while more expression level of agnoprotein was detected, less amount of FEZ1 cosedimented with microtubules and vice-versa. The authors suggested that there might be some conformational change in the C-terminal of FEZ1 when there is an interaction with agnoprotein, resulting in the disruption of FEZ1-microtubule binding. This disruption could also explain the reason why the presence of agnoprotein inhibited the neurite extension in PC12 cells, since FEZ1 and its association with microtubule could lead to neurite outgrowth[37].
Another study with a gene array performed in virus-resistant cell mutant isolated from Rat2 cell, a parental fibroblast line, named R3-2 was conducted in order to investigate the resistant phenotype of an apparent constrain of the viral DNA to the cytoplasm. After confirming the results from microarray through quantitative RT-PCR, FEZ1 gene was up-regulated > 30-fold in the R3-2 cell line. Stable Rat2 cell line over-expressing myc-His-FEZ1 and controls were infected with retroviruses (MLV: Moloney murine leukemia, and HIV-1: human immunodeficiency virus type 1). The cells over-expressing FEZ1 and infected with both viruses showed significant resistance when compared to control cells. This was later confirmed by knockdown of FEZ1 in mutant and wild-type cells. In addition, human cells 293T stably overexpressing FEZ1 and infected with HIV-1 were also capable of blocking infection when compared to control[38].
To assess the time point of this mechanism, the course of viral DNA was analyzed by checking the amount of viral DNA from different intermediates throughout the infection (total viral DNA, earlier steps in reverse transcription and nuclear viral DNA). When compared to R3-2 and controls, cells over-expressing FEZ1 showed similar phenotype: no defect in overall synthesis of viral DNA and similar levels of DNA at early step in reverse transcription. The difference observed was related to the levels of circular viral DNA: Control cell lines showed high levels of it, while low levels were seen in over-expressed FEZ1 and mutant R3-2 cell lines, suggesting that the viral block occurred after reverse transcription but before nuclear entry[38].
Another study further investigated the resistance of neurons expressing high levels of FEZ1 to HIV-1 infection. Also this work interestingly investigated, especially for didactic purposes, the endogenous expression levels of FEZ1, both as mRNA and protein, in different human brain cell lines. Derived neuronal cells (SH-SY5Y) exhibited the highest levels of endogenous expression, while astrocytes (1321N1) and microglia (CHME3) derived cell lines showed significantly lower levels[39].
Similarly to the experiments previously described with R3-2 cells, the authors infected SH-SY5Y, 1321N1 and CHME3 cell lines with HIV-1 and assessed the susceptibility and the intermediates of infection. They found that SH-SY5Y presented the highest resistance to infection. Regarding the total viral DNA, all the three cell lines presented similar levels, while the levels of circular DNA was significantly lower in SH-SY5Y. These experiments were conversely confirmed by RNA interference specific for FEZ1. Both results showed that there was a robust block of HIV-1 infection in the neuronal cell line highly expressing endogenous FEZ1, especially regarding nuclear trafficking of viral DNA[39].
Additional experiments with microglia cell line CHME3 transiently and stably overexpressing FEZ1 were also performed and again showed that the presence of FEZ1 made cells more resistant to viral DNA before nuclear entry. It is noteworthy to mention that FEZ1 expression was not induced in CHME3 by interferon treatment, and as suggested by the authors, the blocking role of FEZ1 may come from its natural expression as a neuron-specific determinant of retroviral infectivity, instead of part of a wider antiviral response[39].
One more study associating FEZ1 with retroviral infection shed light on how this protein affects the delivery of viral DNA to the nucleus. Firstly it was shown that different cells lines presented a potent reduction in HIV-1 infection in the presence of FEZ1 siRNA. Besides CHME3, the cells tested this time were primary normal human dermal fibroblasts (NHDFs) and macrophages (differentiated Thp-1). Conversely, NHDFs cells overexpressing FEZ were able to restore the infection. These results may present a contradiction, however the authors pointed that the effect of FEZ1 as a positive regulator of HIV-1 infection began to decline in cells expressing very high level of this protein, suggesting that indeed the antiviral effect of FEZ1 previously reported may be related to the excessively high expression level of FEZ1 in neuronal cells lines[40].
More strikingly, this work showed that cell extracts containing FEZ1-FLAG could interact to in vitro assembled HIV-1 capsid-nucleocapsid (CA-NC). Moreover, using FEZ1 siRNA and infecting NHDFs with HIV-1 wild-type as well as two HIV-1 mutated capsids (N74D and P90A), both related to entry of the virus into the nucleus, it was reported that the block of infection was similar to all forms of HIV-1. This suggested that FEZ1 was not directly participating in the nuclear entry process but instead could affect the movement of the virus to the nucleus[40].
Since FEZ1 was already reported as a kinesin-1 adaptor, the authors assessed the role of kinesin-1 regarding HIV-1 infection and trafficking. They confirmed that viral particles in knockdown cells for kinesin-1 entered the cytosol and remained largely at the cell periphery, failing to move towards the nucleus. Also using stable NHDFs expressing FEZ1-FLAG and FEZ1-S58A-FLAG (mutant that fails to bind kinesin-1) with further infection with HIV-1, it was reported that only FEZ1-FLAG increased infection, while the mutant did not when compared to control. Therefore it was proposed that FEZ1 binds to HIV-1 and that the virus exploits the role of FEZ1 as a kinesin-1 adaptor, regulating the trafficking of viral particles to the nucleus. Additionally, it was pointed out that while high levels of FEZ1 in neurons may attribute a role as an antiviral factor, in other cells FEZ1 may actually work as a positive host cofactor that facilitates infection by the stated mechanism[40].
The same authors (Malikov and Naghavi[41] in 2017) further provided more details about the role of phosphorylated FEZ1 regarding transport and uncoating of HIV-1. In addition to the association of FEZ1 and Kinesin-1 heavy chain (Kif5B) to regulate virus trafficking, it was also reported that both FEZ1 and Kif5B were required to regulate uncoating in infected siRNA microglia (CHME3) by in situ fluorescence microscopy. In Jurkat cells, FEZ1 also needed to bind Kif5B to promote HIV-1 infection and uncoating[41].
The strategy of HIV-1 to exploit microtubules for intracellular movement was reported to be mediated by FEZ1, since FEZ1 efficiently interacted with HIV-1 core, and no direct interaction between HIV-1 core and Kif5B has been identified. Conversely, the authors reported that Kif5B was expressed in the FEZ1-S58A mutant cells that exhibited defects in transport and uncoating, therefore FEZ1 was suggested to be the bridging factor that enables kinesins to control HIV-1 motility and capsid disassembly[41].
Finally, in line with previous reported work about FEZ1 phosphorylation[20], the authors also demonstrated that in 293T cells over-expressing GFP-MARK2 and FEZ1-FLAG, there was an increase in FEZ1 S58 phosphorylation. In addition, the amounts of HIV-1 CA-NC bound to phosphorylated FEZ1 were also increased. Moreover, the depletion of FEZ1 abrogated the ability of MARK2 expression to enhance infection, thus suggested by the authors that MARK2 effects occurred through its substrate, FEZ1. Therefore, they proposed that HIV-1 binds MARK2 to regulate FEZ1 phosphorylation on HIV-1 cores[41].
The UNC-69 protein in C. elegans is the homolog of mammalian short-coiled coil protein (SCOCO). SCOCO presents a coiled-coil region conserved among species with the N-terminus showed to be more divergent. Conserved function was seen by restoration of locomotion in unc-69 mutants in the presence of human SCOCO. In addition, unc-69 fly mutants showed defective axonal outgrowth and guidance. Northern blot analysis in human fetal tissue showed expression in lung, liver, kidney, and it was enriched in the fetal brain[42].
The interaction with UNC-76 was first identified by yeast two-hybrid screen and subsequently confirmed in vitro by pull-down assay, showing that the coiled-coil of UNC-76 was important for interaction. Moreover, UNC-76 and UNC-69 co-localized in round, perinuclear dots in the soma of C. elegans. This co-localization was disrupted in unc-116 (KHC) mutants, although axonal transport was still occurring. The authors proposed that UNC-69 and UNC-76 could act together in order to promote extension and regulation of synaptic vesicles in axons[42].
Another aspect of SCOCO-FEZ1 interaction is the role in autophagy. Autophagy is a catabolic process and it is initiate by autophagosome formation by activation of UNC-51-like kinase (ULK1)[43,44]. The process requires transport of organelles and macromolecules to the lysosomes for degradation[45-47].
It was previously reported that UNC-51 binds to UNC-76[18]. The mammalian ortholog of UNC-51 is the ULK1 protein and its activation is the initial step of autophagy. Co-IP experiments revealed that ULK1 interacts with FEZ1 and mutations in FEZ1 that disrupted the interaction with SCOCO did not disturbed ULK1-FEZ1 interaction. There was no interaction between ULK1 and SCOCO and siRNA depletion of SCOCO affected FEZ1 and ULK1 interaction. The authors performed a screening in HEK293 cells and found that SCOCO was required for starvation-induced autophagy. They also found that both SCOCO and FEZ1 interacted with UV radiation resistance associated gene (UVRAG), another protein that is also a member of autophagy complexes, by co-IP, and that SCOCO-UVRAG was regulated by FEZ1 and sensitive to starvation[43].
A subsequent study showed that FEZ1 co-localized with RAB3GAP1, which is part of a complex that modulates autophagy[48]. It had been previously reported by yeast two-hybrid screen that these proteins interacted[13]. Spang et al[48] in 2014 showed that the knockdown of FEZ1 and FEZ2 increased autophagic activity, while deficiency of RAB3GAP1/2 decreased this activity.
The autophagy scenario is complex, highly regulated, and the previously related function of FEZ1 as an adaptor of cargo transport - and probably scaffolds protein - places FEZ proteins as emergent targets in this process. Indeed, thirteen FEZ interaction partners are involved in autophagy (Table 2). As already described, the complex of FEZ1 with ULK1, SCOC, RAB3GAP1 or RAB3GAP2 is already correlated to autophagy. However, there is no work demonstrating the involvement of FEZ1 complex with TBC1D25, HAP1, HTT, TLK2, NBR1, PTPRS or TTR in this process.
| Gene | Protein | FEZ1 interaction reference | Autophagy reference |
| RAB3GAP2 | Rab3 GTPase-activating protein non-catalytic subunit | [13] | [48] |
| RAB3GAP1 | Rab3 GTPase-activating protein catalytic subunit | [13] | [48] |
| FEZ1 | Fasciculation and elongation protein zeta-1 | [13] | [43] |
| FEZ2 | Fasciculation and elongation protein zeta-2 | [13] | [43] |
| TBC1D25 | TBC1 domain family member 25 | [13] | [49] |
| HAP1 | Huntingtin-associated protein 1 | [57] | [50] |
| HTT | Huntingtin | [57] | [51] |
| SCOCO | Short coiled-coil protein | [13] | [43] |
| TLK2 | Serine/threonine-protein kinase tousled-like 2 | [13] | [43] |
| NBR1 | Next to BRCA1 gene 1 protein | [6] | [52] |
| PTPRS | PTPRS protein | [55] | [53] |
| TTR | Transthyretin | [55] | [54] |
| ULK1 | UNC-51 like kinase 1 | [43] | [56] |
TBC1D25, also known as OATL1, is a putative Rab guanosine triphosphatase-activating protein (GAP) that interacts with Atg8 homologues and participates in the fusion of lysosomes and autophagosomes[49]. In 2014, it was demonstrated in neurons that HTT and HAP1 are regulators of autophagosome retrograde transport. HAP1 interacts with HTT and control dynein and kinesin motors during the processive transport[50]. Moreover, HTT is able to bind ULK1, releasing ULK1 from negative regulation by mTOR[51]. TLK2, a nuclear kinase, was reported as a negative regulator of autophagy, however the mechanism remains elusive[43]. Several targets are ubiquitinated for degradation and NBR1 is proposed to act as receptor for these targets at autophagosome[52]. PTPRS is a protein tyrosine phosphatase involved in several cell functions and its loss causes autophagy induction[53]. Finally, a mutation in TTR (Y114C) has impaired autophagy[54]. Further studies about FEZ proteins’ involvement in autophagy, if it acts as a protein adaptor or scaffold for proteins above described, can shed light in neurological disorders and cancer[55-57]. It is also important to mention that two studies provided insights on SCOCO’s structure and its binding to FEZ1[58,59].
Another important aspect of FEZ function lies around its transcriptional regulators identified as interactors (Table 3). A general scheme of FEZ protein and its nuclear function is depicted on Figure 6.
| Protein | Description | Reference |
| DRAP1 (NC2α) | DR1 associated protein 1 (negative co-factor 2 alpha) | [13] |
| BAF60a (SMARCD1) | Component of SWI/SNF chromatin remodeling complex | [13] |
| SAP30L | Sin3A-associated protein, 30 kDa-like | [2,13] |
| BRD1 | Bromodomain containing protein 1 | [13] |
| TLK2 | Tousled like kinase 2 | [13] |
| ZNF251 | Zinc finger protein 251 | [13] |
| RARA | Retinoic acid receptor alpha | [2,61] |
| MED7 | Mediator complex subunit 7 | [2] |
| MLF1IP | MLF1 interacting protein | [2] |
| SAP30 | Sin3A-associated protein, 30kDa | [2] |
| SFRS8 | Splicing factor, arginine/serine-rich 8 | [2] |
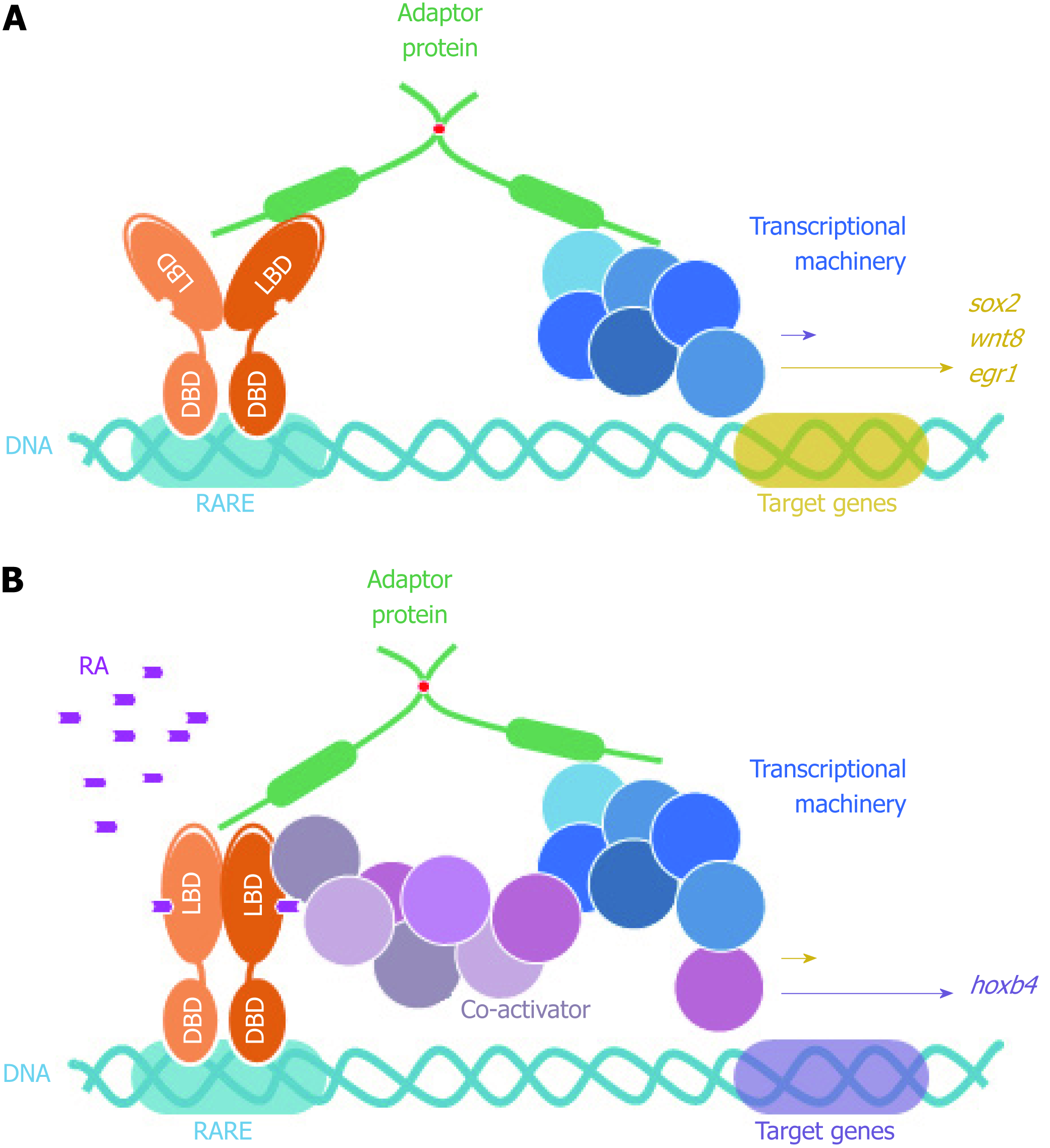
In addition, and still in reference to the permanent SCOCO partner aforementioned, Papanayotou et al[60] in 2008 showed that BERT (Gallus gallus SCOCO), which is equivalent to human SCOCO, participates in the regulation of Sox2 gene expression in the neural plate. This regulation is performed by the binding of BERT to both Geminin and ERNI. Upon BERT binding, the interaction between Geminin-ERNI is disrupted, releasing HP1γ-ERNI-BERT and leaving BERT-Geminin-Brahma in the N2 enhancer and causing the induction of Sox2[60].
In this context, it was recently reported that Sox2 gene was induced when FEZ1 was over-expressed in U87 cells and this was confirmed when FEZ1 was depleted and there was no Sox2 activation, supporting the hypothesis that the presence of FEZ1 could be part of a regulatory complex responsible for Sox2 activation. Moreover, the authors also reported a dramatic induction of Hoxb4 gene in the context of FEZ1 over-expression and the interaction with the retinoic acid receptor (RAR), in the presence of the ligand retinoic acid[61]. Hoxb4 is known to be related to development[62] and experiments showed Fez1 expression by in situ hybridization in chicken embryos during neurulation and somitogenesis (Kobarg et al, non-published observation).
More surprisingly, HOXB4 was reported to correlate with acute myeloid leukemia[63], which is very consistent with our findings regarding increased expression of FEZ1 in this disease by immunohistochemistry experiments with tissues from patients (Kobarg et al, not published). The interesting link between FEZ1 and HOXB4 differential expression in acute myeloid leukemia could lie in the discovery that FEZ1 over-expression caused the phenotype of multi lobulated nuclei (also known as flower-like nuclei) in mammalian cell line[64]. This nuclear phenotype has already been reported to be a marker of myeloid leukemia of M4/5 subtype[65,66].
It is important to mention that another unrelated protein had been synonymously called “FEZ1”/LZTS1 (Q9Y250, 596 amino acids length), in the past. This transcription factor and tumor suppressor protein has been reported also as “FEZ1” (“F37 /Esophageal cancer-related gene-coding leucine Zipper motif 1”)[67] but today has been renamed to: LZTS1 (“Leucine zipper putative Tumor Suppressor 1”), and is no longer called FEZ1[68].
FEZ proteins are implicated in neuronal development and viral infection and their role as kinesin adaptor has been studied in these context. However, FEZ proteins are hubs and novel nuclear functions that could be involved in these processes have been described. The structural, evolutionary and functional data available for this family of proteins provides tools to understand the role of hub proteins in physiological and pathological states.
| 1. | Bloom L, Horvitz HR. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc Natl Acad Sci USA. 1997;94:3414-3419. [PubMed] |
| 2. | Alborghetti MR, Furlan AS, Kobarg J. FEZ2 has acquired additional protein interaction partners relative to FEZ1: functional and evolutionary implications. PLoS One. 2011;6:e17426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1363] [Cited by in RCA: 1226] [Article Influence: 68.1] [Reference Citation Analysis (0)] |
| 4. | Honda A, Miyoshi K, Baba K, Taniguchi M, Koyama Y, Kuroda S, Katayama T, Tohyama M. Expression of fasciculation and elongation protein zeta-1 (FEZ1) in the developing rat brain. Brain Res Mol Brain Res. 2004;122:89-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Fujita T, Ikuta J, Hamada J, Okajima T, Tatematsu K, Tanizawa K, Kuroda S. Identification of a tissue-non-specific homologue of axonal fasciculation and elongation protein zeta-1. Biochem Biophys Res Commun. 2004;313:738-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Whitehouse C, Chambers J, Howe K, Cobourne M, Sharpe P, Solomon E. NBR1 interacts with fasciculation and elongation protein zeta-1 (FEZ1) and calcium and integrin binding protein (CIB) and shows developmentally restricted expression in the neural tube. Eur J Biochem. 2002;269:538-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 7. | He J, Liu J, Zhang Z, Sun M, Zhu T, Xia C. Expression of fasciculation and elongation protein zeta-1 (FEZ1) in cultured rat neonatal astrocytes. Mol Cell Biochem. 2009;325:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 8. | Sun T, Yu N, Zhai LK, Li N, Zhang C, Zhou L, Huang Z, Jiang XY, Shen Y, Chen ZY. c-Jun NH2-terminal kinase (JNK)-interacting protein-3 (JIP3) regulates neuronal axon elongation in a kinesin- and JNK-dependent manner. J Biol Chem. 2013;288:14531-14543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Chen X, Ku L, Mei R, Liu G, Xu C, Wen Z, Zhao X, Wang F, Xiao L, Feng Y. Novel schizophrenia risk factor pathways regulate FEZ1 to advance oligodendroglia development. Transl Psychiatry. 2017;7:1293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Lanza DC, Silva JC, Assmann EM, Quaresma AJ, Bressan GC, Torriani IL, Kobarg J. Human FEZ1 has characteristics of a natively unfolded protein and dimerizes in solution. Proteins. 2009;74:104-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Alborghetti MR, Furlan AS, Silva JC, Paes Leme AF, Torriani IC, Kobarg J. Human FEZ1 protein forms a disulfide bond mediated dimer: implications for cargo transport. J Proteome Res. 2010;9:4595-4603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Makino T, Suzuki Y, Gojobori T. Differential evolutionary rates of duplicated genes in protein interaction network. Gene. 2006;385:57-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Assmann EM, Alborghetti MR, Camargo ME, Kobarg J. FEZ1 dimerization and interaction with transcription regulatory proteins involves its coiled-coil region. J Biol Chem. 2006;281:9869-9881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Gindhart JG, Chen J, Faulkner M, Gandhi R, Doerner K, Wisniewski T, Nandlestadt A. The kinesin-associated protein UNC-76 is required for axonal transport in the Drosophila nervous system. Mol Biol Cell. 2003;14:3356-3365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 73] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 15. | Sakae N, Yamasaki N, Kitaichi K, Fukuda T, Yamada M, Yoshikawa H, Hiranita T, Tatsumi Y, Kira J, Yamamoto T, Miyakawa T, Nakayama KI. Mice lacking the schizophrenia-associated protein FEZ1 manifest hyperactivity and enhanced responsiveness to psychostimulants. Hum Mol Genet. 2008;17:3191-3203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 16. | Sumitomo A, Saka A, Ueta K, Horike K, Hirai K, Gamo NJ, Hikida T, Nakayama KI, Sawa A, Sakurai T, Tomoda T. Methylphenidate and Guanfacine Ameliorate ADHD-Like Phenotypes in Fez1-Deficient Mice. Mol Neuropsychiatry. 2018;3:223-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Kuroda S, Nakagawa N, Tokunaga C, Tatematsu K, Tanizawa K. Mammalian homologue of the Caenorhabditis elegans UNC-76 protein involved in axonal outgrowth is a protein kinase C zeta-interacting protein. J Cell Biol. 1999;144:403-411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Toda H, Mochizuki H, Flores R, Josowitz R, Krasieva TB, Lamorte VJ, Suzuki E, Gindhart JG, Furukubo-Tokunaga K, Tomoda T. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 2008;22:3292-3307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 104] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 19. | Chua JJ, Butkevich E, Worseck JM, Kittelmann M, Grønborg M, Behrmann E, Stelzl U, Pavlos NJ, Lalowski MM, Eimer S, Wanker EE, Klopfenstein DR, Jahn R. Phosphorylation-regulated axonal dependent transport of syntaxin 1 is mediated by a Kinesin-1 adapter. Proc Natl Acad Sci USA. 2012;109:5862-5867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Butkevich E, Härtig W, Nikolov M, Erck C, Grosche J, Urlaub H, Schmidt CF, Klopfenstein DR, Chua JJ. Phosphorylation of FEZ1 by Microtubule Affinity Regulating Kinases regulates its function in presynaptic protein trafficking. Sci Rep. 2016;6:26965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Okumura F, Hatakeyama S, Matsumoto M, Kamura T, Nakayama KI. Functional regulation of FEZ1 by the U-box-type ubiquitin ligase E4B contributes to neuritogenesis. J Biol Chem. 2004;279:53533-53543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 22. | Lee S, Walker CL, Karten B, Kuny SL, Tennese AA, O'Neill MA, Wevrick R. Essential role for the Prader-Willi syndrome protein necdin in axonal outgrowth. Hum Mol Genet. 2005;14:627-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 98] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 23. | Watanabe Y, Khodosevich K, Monyer H. Dendrite development regulated by the schizophrenia-associated gene FEZ1 involves the ubiquitin proteasome system. Cell Rep. 2014;7:552-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Bryant DT, Landles C, Papadopoulou AS, Benjamin AC, Duckworth JK, Rosahl T, Benn CL, Bates GP. Disruption to schizophrenia-associated gene Fez1 in the hippocampus of HDAC11 knockout mice. Sci Rep. 2017;7:11900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Fujita T, Maturana AD, Ikuta J, Hamada J, Walchli S, Suzuki T, Sawa H, Wooten MW, Okajima T, Tatematsu K, Tanizawa K, Kuroda S. Axonal guidance protein FEZ1 associates with tubulin and kinesin motor protein to transport mitochondria in neurites of NGF-stimulated PC12 cells. Biochem Biophys Res Commun. 2007;361:605-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 55] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Ikuta J, Maturana A, Fujita T, Okajima T, Tatematsu K, Tanizawa K, Kuroda S. Fasciculation and elongation protein zeta-1 (FEZ1) participates in the polarization of hippocampal neuron by controlling the mitochondrial motility. Biochem Biophys Res Commun. 2007;353:127-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Sure GR, Chatterjee A, Mishra N, Sabharwal V, Devireddy S, Awasthi A, Mohan S, Koushika SP. UNC-16/JIP3 and UNC-76/FEZ1 limit the density of mitochondria in C. elegans neurons by maintaining the balance of anterograde and retrograde mitochondrial transport. Sci Rep. 2018;8:8938. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 28. | Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ. Two binding partners cooperate to activate the molecular motor Kinesin-1. J Cell Biol. 2007;176:11-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 195] [Cited by in RCA: 179] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 29. | Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T, Kuroda S, Katayama T, Tohyama M. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 2003;8:685-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 247] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 30. | Kang E, Burdick KE, Kim JY, Duan X, Guo JU, Sailor KA, Jung DE, Ganesan S, Choi S, Pradhan D, Lu B, Avramopoulos D, Christian K, Malhotra AK, Song H, Ming GL. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72:559-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 31. | Yamada K, Nakamura K, Minabe Y, Iwayama-Shigeno Y, Takao H, Toyota T, Hattori E, Takei N, Sekine Y, Suzuki K, Iwata Y, Miyoshi K, Honda A, Baba K, Katayama T, Tohyama M, Mori N, Yoshikawa T. Association analysis of FEZ1 variants with schizophrenia in Japanese cohorts. Biol Psychiatry. 2004;56:683-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Koga M, Ishiguro H, Horiuchi Y, Albalushi T, Inada T, Iwata N, Ozaki N, Ujike H, Muratake T, Someya T, Arinami T. Failure to confirm the association between the FEZ1 gene and schizophrenia in a Japanese population. Neurosci Lett. 2007;417:326-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 33. | Hodgkinson CA, Goldman D, Ducci F, DeRosse P, Caycedo DA, Newman ER, Kane JM, Roy A, Malhotra AK. The FEZ1 gene shows no association to schizophrenia in Caucasian or African American populations. Neuropsychopharmacology. 2007;32:190-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 34. | Lipska BK, Mitkus SN, Mathew SV, Fatula R, Hyde TM, Weinberger DR, Kleinman JE. Functional genomics in postmortem human brain: abnormalities in a DISC1 molecular pathway in schizophrenia. Dialogues Clin Neurosci. 2006;8:353-357. [PubMed] |
| 35. | Vachev TI, Stoyanova VK, Ivanov HY, Minkov IN, Popov NT. Investigation of fasciculation and elongation protein ζ-1 (FEZ1) in peripheral blood reveals differences in gene expression in patients with schizophrenia. Balkan J Med Genet. 2015;18:31-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 36. | Colantuoni C, Hyde TM, Mitkus S, Joseph A, Sartorius L, Aguirre C, Creswell J, Johnson E, Deep-Soboslay A, Herman MM, Lipska BK, Weinberger DR, Kleinman JE. Age-related changes in the expression of schizophrenia susceptibility genes in the human prefrontal cortex. Brain Struct Funct. 2008;213:255-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 37. | Suzuki T, Okada Y, Semba S, Orba Y, Yamanouchi S, Endo S, Tanaka S, Fujita T, Kuroda S, Nagashima K, Sawa H. Identification of FEZ1 as a protein that interacts with JC virus agnoprotein and microtubules: role of agnoprotein-induced dissociation of FEZ1 from microtubules in viral propagation. J Biol Chem. 2005;280:24948-24956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 38. | Naghavi MH, Hatziioannou T, Gao G, Goff SP. Overexpression of fasciculation and elongation protein zeta-1 (FEZ1) induces a post-entry block to retroviruses in cultured cells. Genes Dev. 2005;19:1105-1115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 39. | Haedicke J, Brown C, Naghavi MH. The brain-specific factor FEZ1 is a determinant of neuronal susceptibility to HIV-1 infection. Proc Natl Acad Sci USA. 2009;106:14040-14045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 40. | Malikov V, da Silva ES, Jovasevic V, Bennett G, de Souza Aranha Vieira DA, Schulte B, Diaz-Griffero F, Walsh D, Naghavi MH. HIV-1 capsids bind and exploit the kinesin-1 adaptor FEZ1 for inward movement to the nucleus. Nat Commun. 2015;6:6660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 41. | Malikov V, Naghavi MH. Localized Phosphorylation of a Kinesin-1 Adaptor by a Capsid-Associated Kinase Regulates HIV-1 Motility and Uncoating. Cell Rep. 2017;20:2792-2799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 42. | Su CW, Tharin S, Jin Y, Wightman B, Spector M, Meili D, Tsung N, Rhiner C, Bourikas D, Stoeckli E, Garriga G, Horvitz HR, Hengartner MO. The short coiled-coil domain-containing protein UNC-69 cooperates with UNC-76 to regulate axonal outgrowth and normal presynaptic organization in Caenorhabditis elegans. J Biol. 2006;5:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 43. | McKnight NC, Jefferies HB, Alemu EA, Saunders RE, Howell M, Johansen T, Tooze SA. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J. 2012;31:1931-1946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 44. | Chan EY, Tooze SA. Evolution of Atg1 function and regulation. Autophagy. 2009;5:758-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 45. | Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1556] [Cited by in RCA: 1681] [Article Influence: 88.5] [Reference Citation Analysis (0)] |
| 46. | Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5765] [Cited by in RCA: 5824] [Article Influence: 323.6] [Reference Citation Analysis (12)] |
| 47. | Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1253] [Cited by in RCA: 1401] [Article Influence: 93.4] [Reference Citation Analysis (0)] |
| 48. | Spang N, Feldmann A, Huesmann H, Bekbulat F, Schmitt V, Hiebel C, Koziollek-Drechsler I, Clement AM, Moosmann B, Jung J, Behrends C, Dikic I, Kern A, Behl C. RAB3GAP1 and RAB3GAP2 modulate basal and rapamycin-induced autophagy. Autophagy. 2014;10:2297-2309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 49. | Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol. 2011;192:839-853. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 139] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 50. | Wong YC, Holzbaur EL. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci. 2014;34:1293-1305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 309] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 51. | Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, Cuervo AM, Zhang S. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015;17:262-275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 338] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 52. | Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Øvervatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 791] [Cited by in RCA: 917] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 53. | Martin KR, Xu Y, Looyenga BD, Davis RJ, Wu CL, Tremblay ML, Xu HE, MacKeigan JP. Identification of PTPsigma as an autophagic phosphatase. J Cell Sci. 2011;124:812-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 54. | Li H, Zhang Y, Cao L, Xiong R, Zhang B, Wu L, Zhao Z, Chen SD. Curcumin could reduce the monomer of TTR with Tyr114Cys mutation via autophagy in cell model of familial amyloid polyneuropathy. Drug Des Devel Ther. 2014;8:2121-2128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 55. | Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1772] [Cited by in RCA: 1720] [Article Influence: 81.9] [Reference Citation Analysis (1)] |
| 56. | Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464-25474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 373] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 57. | Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, Herbst M, Suopanki J, Scherzinger E, Abraham C, Bauer B, Hasenbank R, Fritzsche A, Ludewig AH, Büssow K, Coleman SH, Gutekunst CA, Landwehrmeyer BG, Lehrach H, Wanker EE. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol Cell. 2004;15:853-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 330] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 58. | Alborghetti MR, Furlan Ada S, da Silva JC, Sforça ML, Honorato RV, Granato DC, dos Santos Migueleti DL, Neves JL, de Oliveira PS, Paes-Leme AF, Zeri AC, de Torriani IC, Kobarg J. Structural analysis of intermolecular interactions in the kinesin adaptor complex fasciculation and elongation protein zeta 1/short coiled-coil protein (FEZ1/SCOCO). PLoS One. 2013;8:e76602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 59. | Behrens C, Binotti B, Schmidt C, Robinson CV, Chua JJ, Kühnel K. Crystal structure of the human short coiled coil protein and insights into SCOC-FEZ1 complex formation. PLoS One. 2013;8:e76355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 60. | Papanayotou C, Mey A, Birot AM, Saka Y, Boast S, Smith JC, Samarut J, Stern CD. A mechanism regulating the onset of Sox2 expression in the embryonic neural plate. PLoS Biol. 2008;6:e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 89] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 61. | Bertini Teixeira M, Figueira ACM, Furlan AS, Aquino B, Alborghetti MR, Paes Leme AF, Wei LN, Kobarg J. Fasciculation and elongation zeta-1 protein (FEZ1) interacts with the retinoic acid receptor and participates in transcriptional regulation of the Hoxb4 gene. FEBS Open Bio. 2018;8:4-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 62. | Garcia-Fernàndez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 340] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 63. | Umeda S, Yamamoto K, Murayama T, Hidaka M, Kurata M, Ohshima T, Suzuki S, Sugawara E, Kawano F, Kitagawa M. Prognostic significance of HOXB4 in de novo acute myeloid leukemia. Hematology. 2012;17:125-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 64. | Lanza DC, Trindade DM, Assmann EM, Kobarg J. Over-expression of GFP-FEZ1 causes generation of multi-lobulated nuclei mediated by microtubules in HEK293 cells. Exp Cell Res. 2008;314:2028-2039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 65. | Shimoyama M, Kagami Y, Shimotohno K, Miwa M, Minato K, Tobinai K, Suemasu K, Sugimura T. Adult T-cell leukemia/lymphoma not associated with human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1986;83:4524-4528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 66. | Graham RL, Burch M, Krause JR. Adult T-cell leukemia/lymphoma. Proc (Bayl Univ Med Cent). 2014;27:235-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 55] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 67. | Ishii H, Baffa R, Numata SI, Murakumo Y, Rattan S, Inoue H, Mori M, Fidanza V, Alder H, Croce CM. The FEZ1 gene at chromosome 8p22 encodes a leucine-zipper protein, and its expression is altered in multiple human tumors. Proc Natl Acad Sci USA. 1999;96:3928-3933. [PubMed] |
| 68. | Lovat F, Ishii H, Schiappacassi M, Fassan M, Barbareschi M, Galligioni E, Gasparini P, Baldassarre G, Croce CM, Vecchione A. LZTS1 downregulation confers paclitaxel resistance and is associated with worse prognosis in breast cancer. Oncotarget. 2014;5:970-977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited manuscript
Specialty type: Biochemistry and molecular biology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Moneim AA, Pierzchalski P S- Editor: Ji FF L- Editor: A E- Editor: Song H