©2011 Baishideng Publishing Group Co.
World J Biol Chem. Mar 26, 2011; 2(3): 48-58
Published online Mar 26, 2011. doi: 10.4331/wjbc.v2.i3.48
Published online Mar 26, 2011. doi: 10.4331/wjbc.v2.i3.48
Figure 1 André Luis Souza dos Santos, Associate Professor, Laboratory of Multidisciplinary Studies on Microbial Biochemistry, Department of General Microbiology, Institute of Microbiology Prof.
Paulo de Góes, Federal University of Rio de Janeiro, Rio de Janeiro, RJ 21941-902, Brazil.
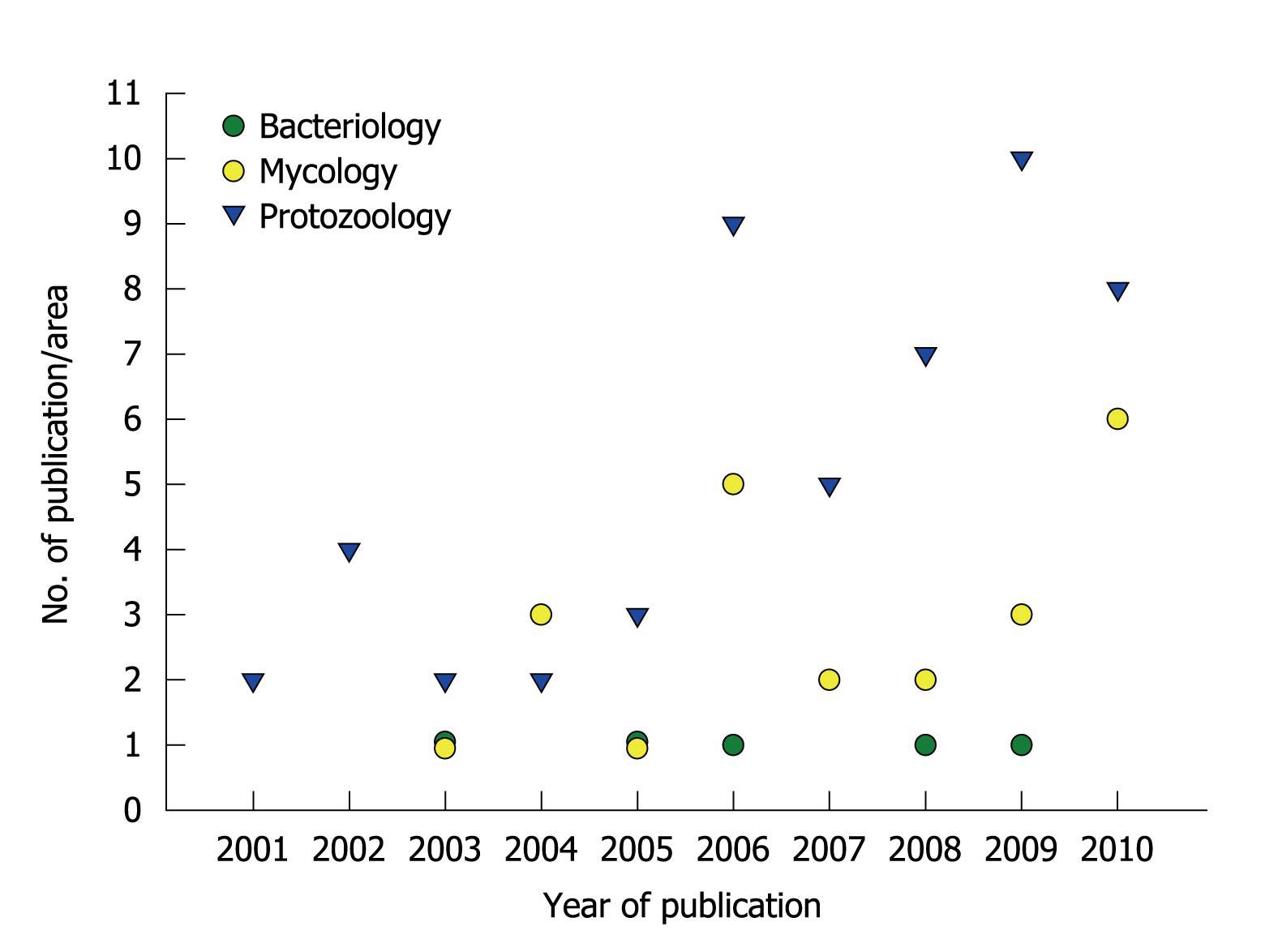
Figure 2 Publication of scientific papers by the research group led by André Santos.
The graphic summarizes the numbers and specific areas of Microbiology in relation to papers published during the past ten years.
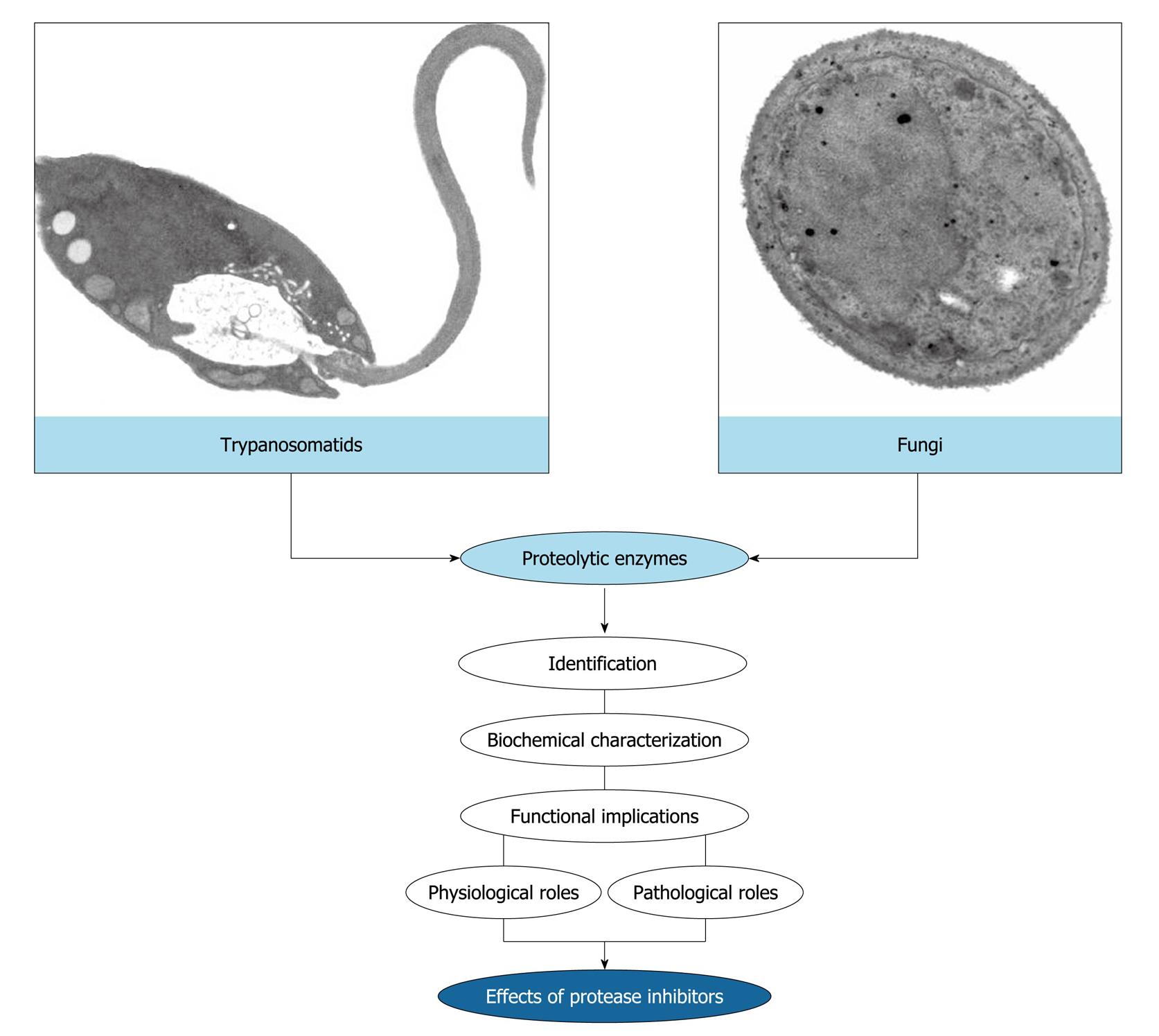
Figure 3 Rationale of the research works developed in the André Santos’ laboratory.
The main purpose of our study focuses on the identification and biochemical characterization of cellular and/or extracellular proteases produced by eukaryotic microorganisms, especially trypanosomatids and fungi. Subsequently, we have focused on the discovery of possible biological functions for these hydrolytic enzymes in both the social context of the microbial cell and the participation in interaction events with biotic and abiotic substrates. Finally, we have used the protease inhibitors in an attempt to block vital processes in microbial cells, thus preventing a successful infection.
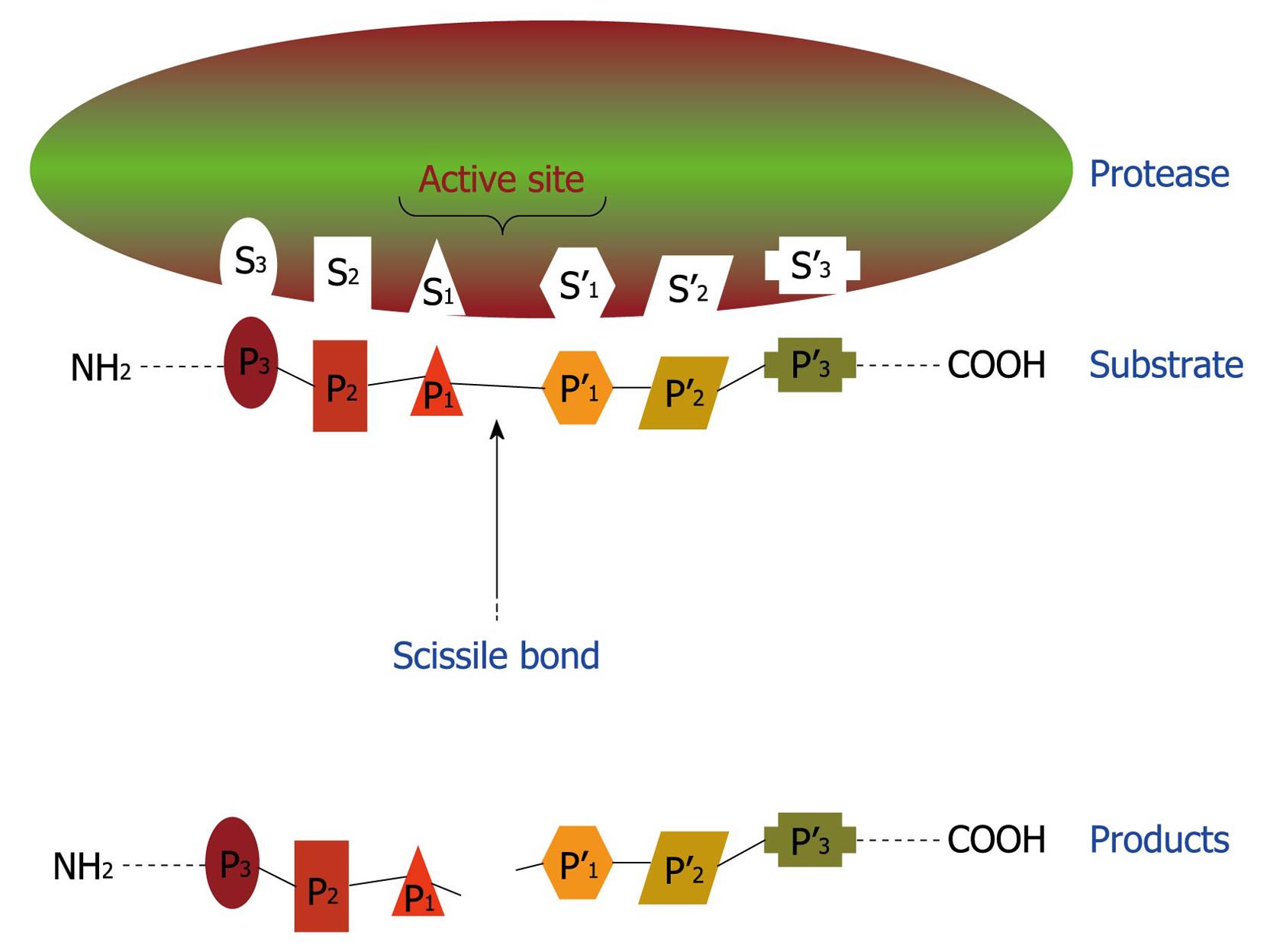
Figure 4 Schematic representation of binding region and catalytic site of a protease.
This hypothetical protease possesses six subsites (S1-S3 and S1’-S3’) in its catalytic site and, consequently, is able to recognize and bind to a sequence of six amino acids (P1-P3 and P1’-P3’) in the proteinaceous substrate. After proteolysis, at least two smaller peptides are generated as the reaction products.
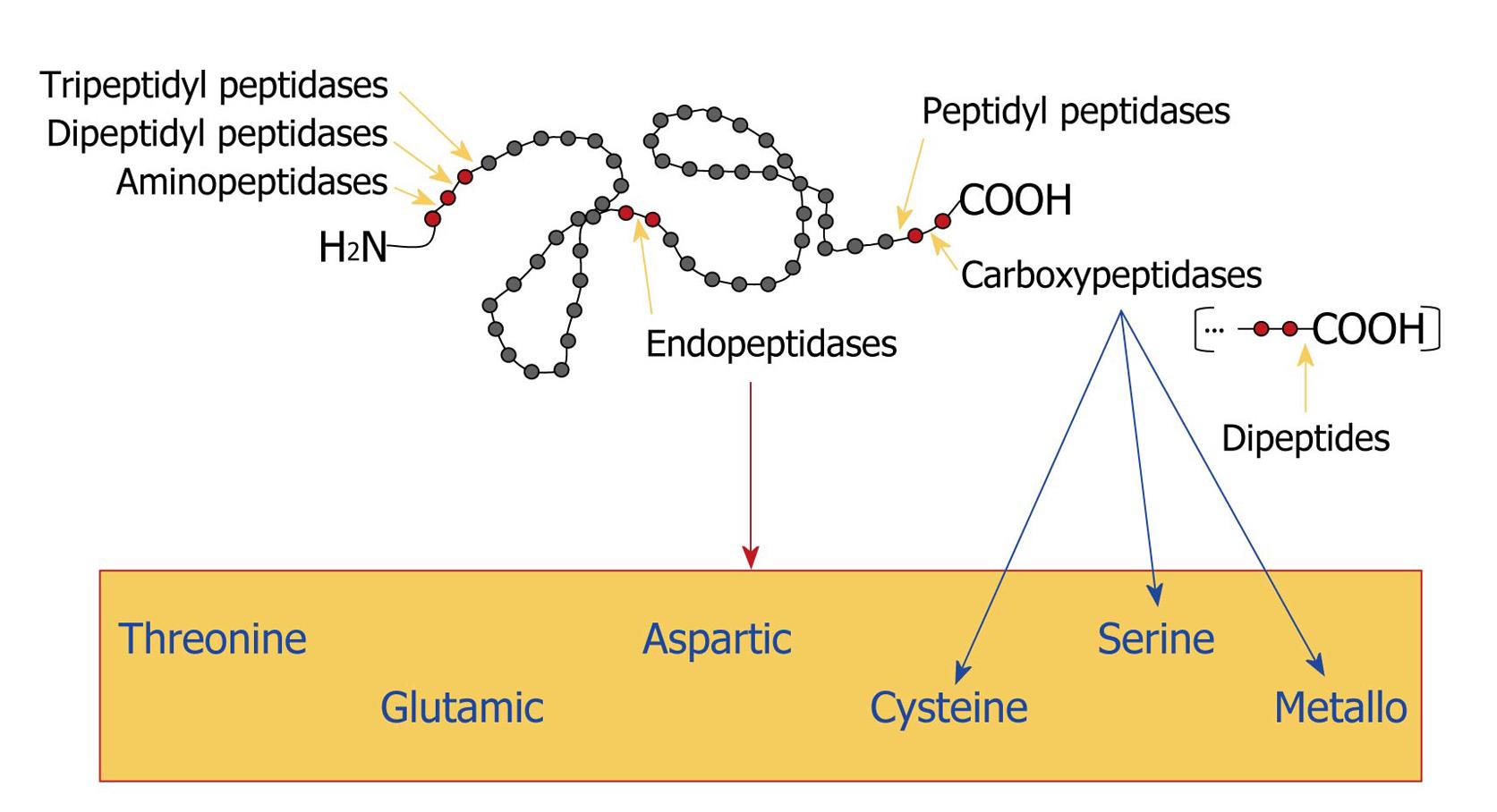
Figure 5 Classification of proteases.
Gray circles represent amino acids and red circles indicate the amino acid sequence that is bound to the proteolytic enzyme. Yellow arrows point to the site of cleavage. The blue arrows indicate the classification of carboxypeptidases and the red arrow shows the box containing all the classes of endopeptidases, according to the chemical group present in their catalytic sites.
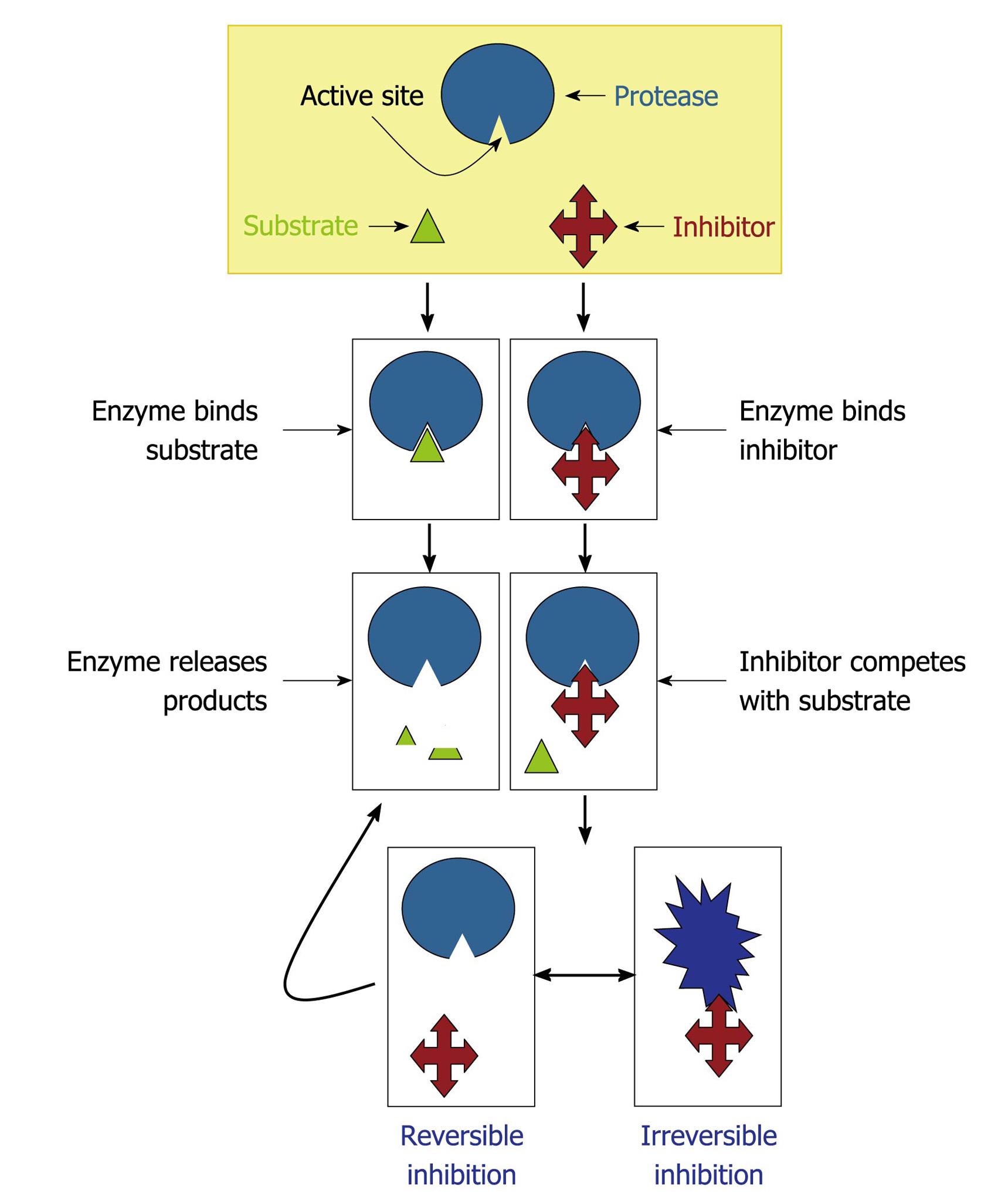
Figure 6 Mechanisms of protease inhibition.
The protease inhibitor competes with the substrate to bind to the active site of a protease and two distinct possibilities arise: (1) substrate binds to the catalytic site and then is cleaved by the protease, which releases the products or (2) inhibitor binds to the active site and by steric hindrance blocks the substrate attachment. In this last case, the inhibitor can promote an irreversible (the conformational structure of the protease is completely lost) or reversible inhibition (when the inhibitor disconnects from the enzyme, the substrate can bind to it).
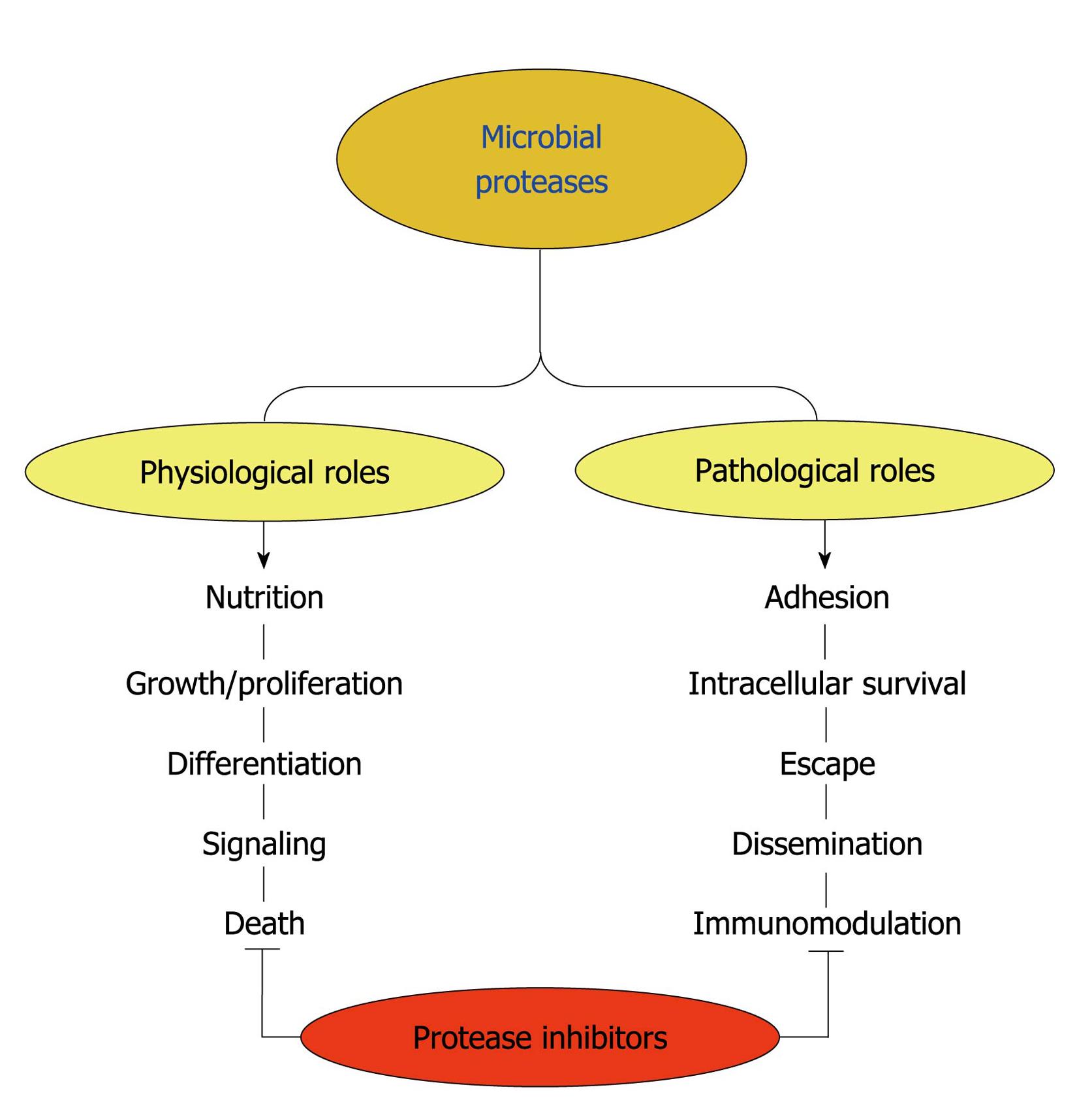
Figure 7 Possible functions played by microbial proteases.
Surface and/or secreted proteases are able to cleave different host components such as serum proteins, antimicrobial peptides, surface molecules and structural proteinaceous compounds. The degradation of host proteins can help the microorganisms in several steps of their life cycle and pathogenesis including dissemination, adhesion, escape, nutrition and immunomodulation of the host immune response. These proteases can also contribute to maintaining basic metabolic processes in a microbial cell, which govern crucial events like proliferation, differentiation, signaling and death pathways. Proteolytic inhibitors are able to block one or several of these fundamental events.
- Citation: Santos ALSD. Protease expression by microorganisms and its relevance to crucial physiological/pathological events. World J Biol Chem 2011; 2(3): 48-58
- URL: https://www.wjgnet.com/1949-8454/full/v2/i3/48.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v2.i3.48