©The Author(s) 2019.
World J Biol Chem. Feb 21, 2019; 10(2): 28-43
Published online Feb 21, 2019. doi: 10.4331/wjbc.v10.i2.28
Published online Feb 21, 2019. doi: 10.4331/wjbc.v10.i2.28
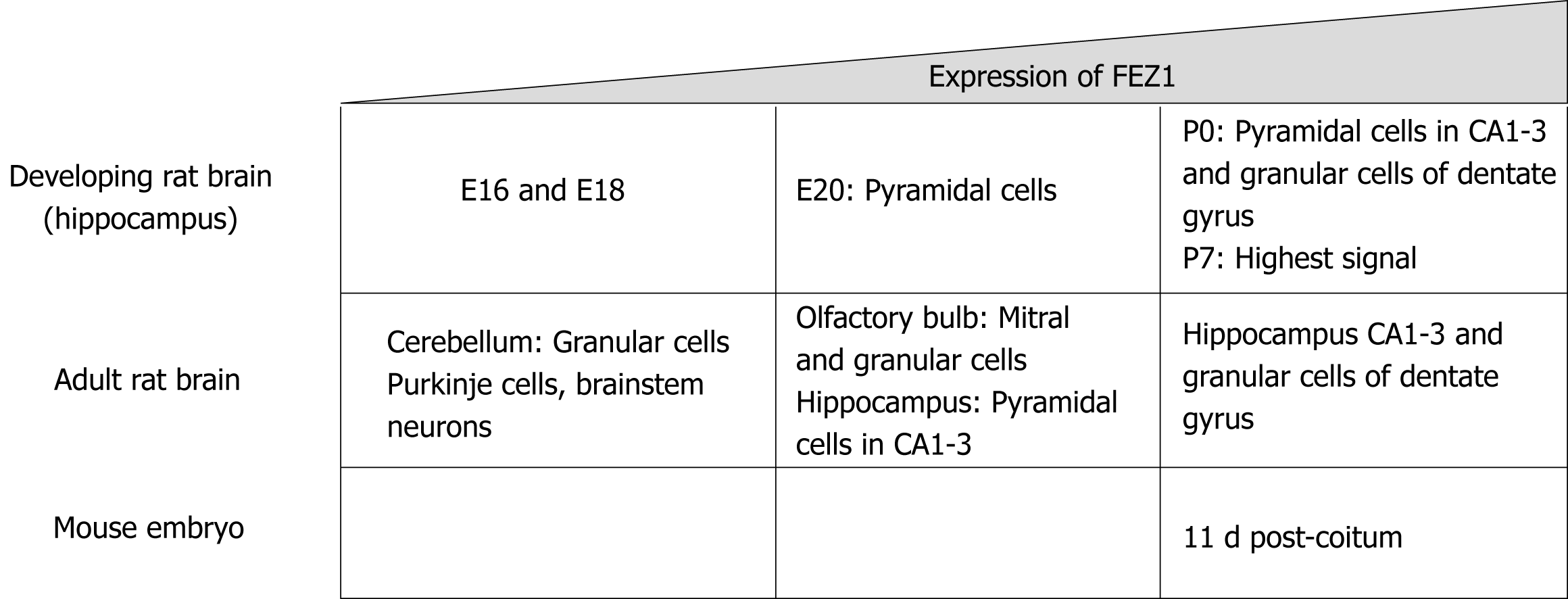
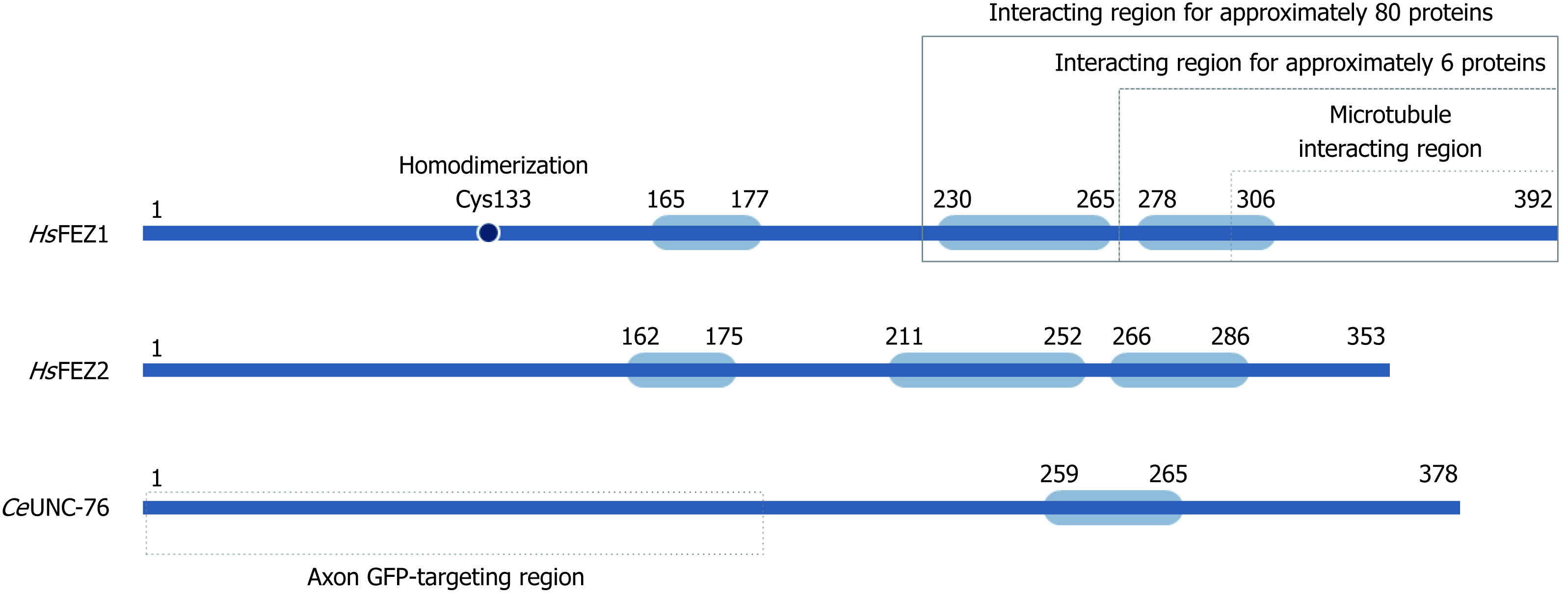
Figure 2 Schematic structure for FEZ proteins.
Blue boxes indicate predicted coiled-coil regions involved in homodimerization or protein-protein interaction.
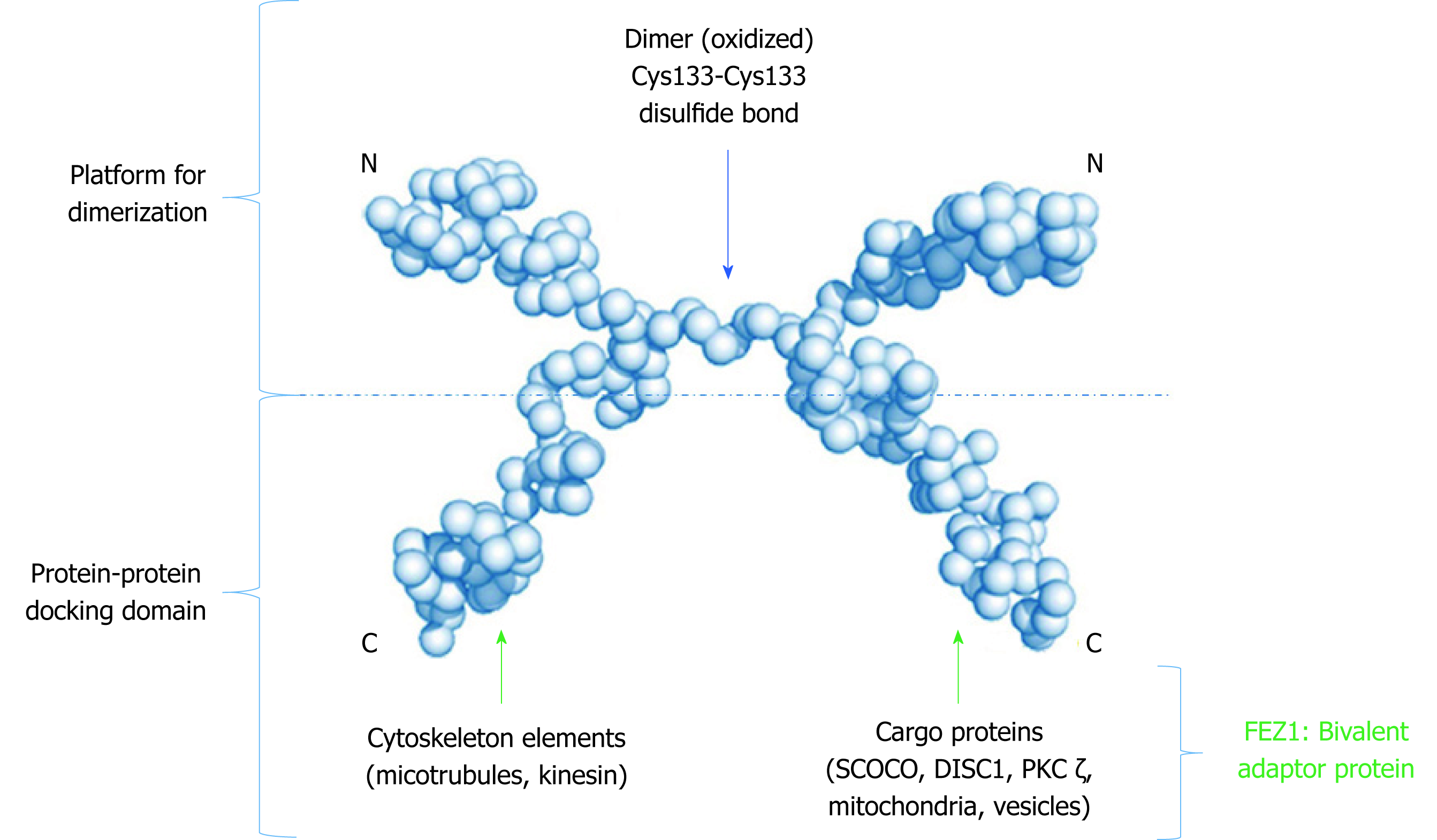
Figure 3 Low resolution model proposed to the dimeric structure of FEZ1.
Monomers connect through a cysteine present in the N-terminal. The C-terminal of each monomer is free to perform protein-protein interactions. Adapted from the model proposed by Alborghetti et al[11], 2010. SCOCO: Short-coiled coil protein; DISC1: Disrupted-In-Schizophrenia 1; PKC: Protein kinase C.
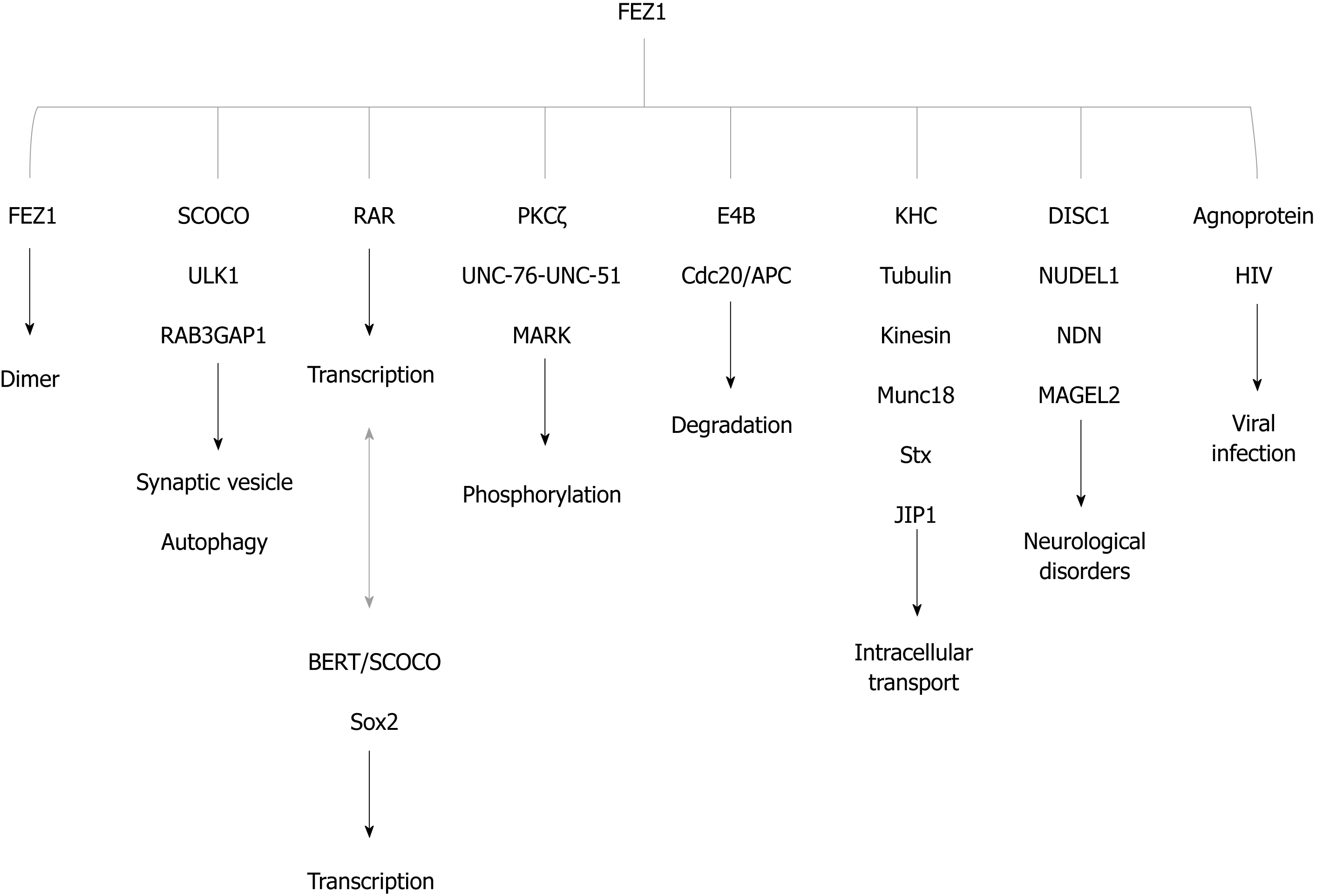
Figure 4 FEZ1 and proteins reported to associate either by direct binding or by the presence in a protein complex.
The function of the association is also depicted. BERT/SCOCO was reported to regulate the expression of Sox2 gene. SCOCO: Short-coiled coil protein; DISC1: Disrupted-In-Schizophrenia 1; PKC: Protein kinase C; KHC: Kinesin heavy chain; RAR: Retinoic acid receptor; HIV: Human immunodeficiency virus; NUDEL: Nuclear distribution element-like; NDN: Necdin; ULK1: UNC-51-like kinase; JIP1: c-Jun N-terminal kinase-interacting protein 1; Stx: Syntaxin 1a; MARK: Microtubule-affinity regulating kinase.
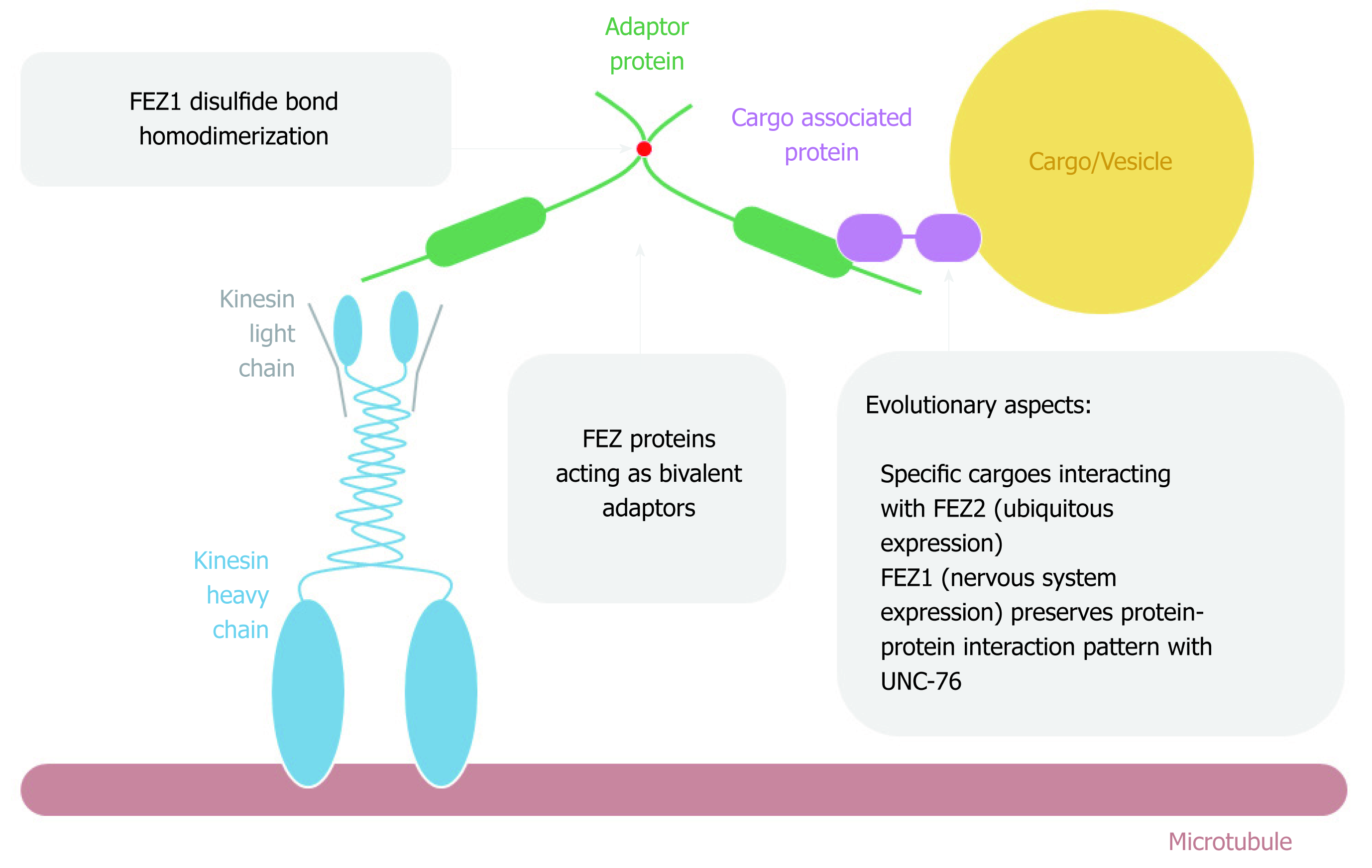
Figure 5 FEZ proteins cytoplasmic function as transport bivalent adaptors.
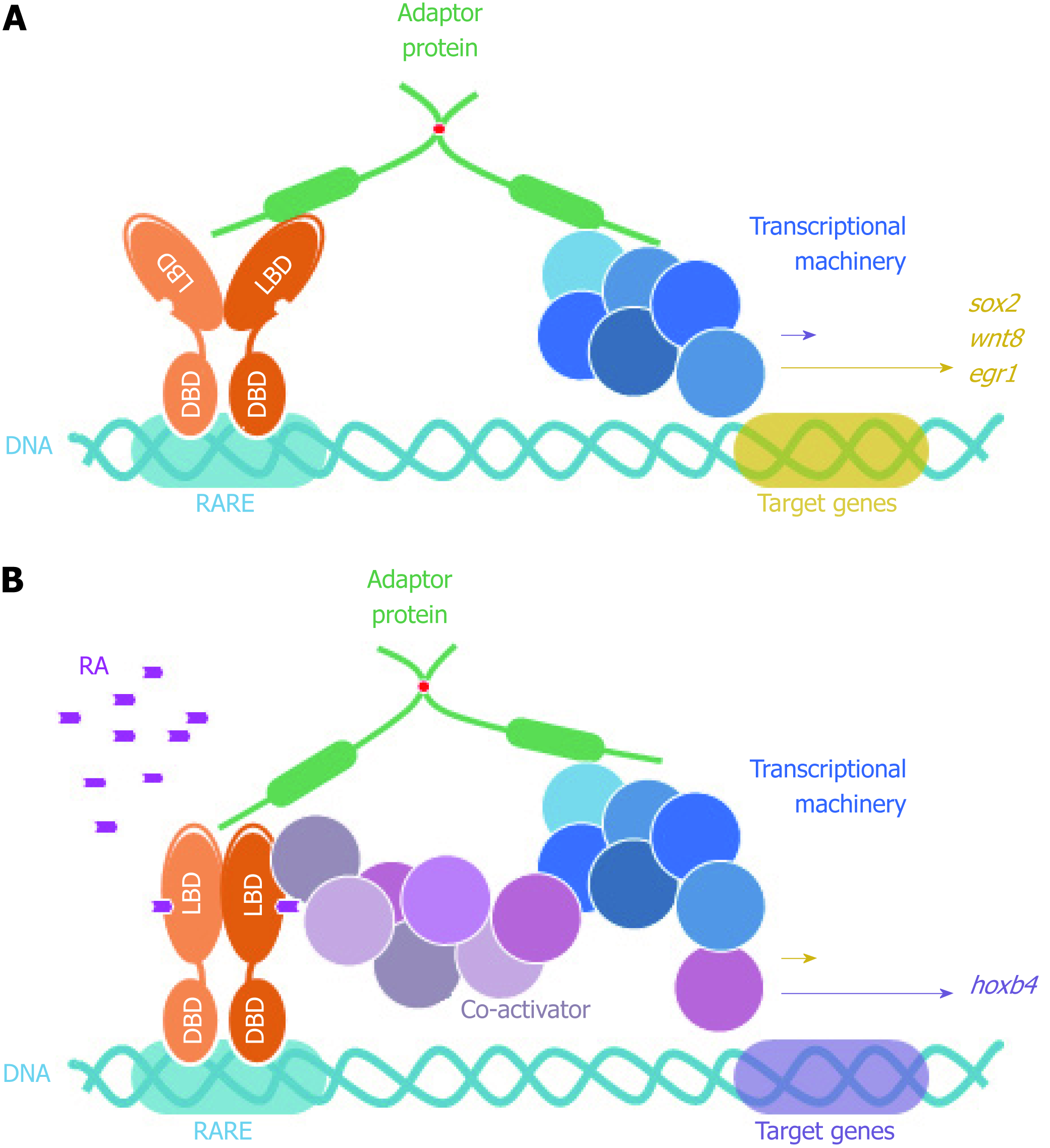
Figure 6 FEZ proteins nuclear function as scaffold between nuclear receptors (retinoic acid receptor) and transcriptional machinery.
DBD: DNA binding domain; LBD: Ligand binding domain; RARE: Retinoic acid response element; RA: Retinoic acid.
- Citation: Teixeira MB, Alborghetti MR, Kobarg J. Fasciculation and elongation zeta proteins 1 and 2: From structural flexibility to functional diversity. World J Biol Chem 2019; 10(2): 28-43
- URL: https://www.wjgnet.com/1949-8454/full/v10/i2/28.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v10.i2.28