Published online Sep 27, 2025. doi: 10.4240/wjgs.v17.i9.109521
Revised: June 4, 2025
Accepted: July 18, 2025
Published online: September 27, 2025
Processing time: 133 Days and 20 Hours
Gallstone (GS), a prevalent biliary disorder, is associated with bile stasis, infection, and cholesterol metabolism. Recent research highlights the potential role of bile microbiota in GS pathogenesis. This is a case control study conducted at Jinshan Hospital between 2022 and 2023.
To investigate the relationship between bile microbiota dysbiosis and GS forma
This is a retrospective analysis conducted at Jinshan Hospital between 2022 and 2023. A total of 40 patients were analyzed, including 25 with GS and 15 with GS-free (GSF). Bile samples from 27 patients were analyzed using 16S rRNA gene sequencing to assess microbial composition.
Significant differences were found in bile acid profiles between GS and GSF groups, with lower microbial diversity in GS patients, indicated by reduced Shan
Dysbiosis, particularly overgrowth of Proteobacteria, may contribute to gallstone formation, while Lactobacillus could play a protective role. Further research is needed to validate these findings.
Core Tip: This retrospective study analyzed bile microbiota composition in 40 patients. Results demonstrated that samples from gallstone patients exhibited significantly reduced overall microbial diversity alongside a striking dominance of Proteobacteria. In contrast, bile samples from gallstone-free individuals showed relative enrichment in health-associated genera, including protective taxa like Lactobacillus. The findings indicate that dysbiosis within the bile microbiome, particularly the overgrowth of Proteobacteria, actively drives gallstone pathogenesis. This suggests that modulating the bile microbiota represents a promising novel therapeutic avenue for gallstone prevention or treatment through targeted interventions.
- Citation: Lu ZX, Jiang YQ, Wang DS, Song YT, Jiang XM, Xu FJ, Tang J, Li B, Huang WH. Gallstone and bile microbiota: A case-control study based on 16S rRNA gene sequencing. World J Gastrointest Surg 2025; 17(9): 109521
- URL: https://www.wjgnet.com/1948-9366/full/v17/i9/109521.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v17.i9.109521
Gallstones (GSs), or cholelithiasis, are a common hepatobiliary disorder caused by abnormal elevations in cholesterol or bilirubin levels in the bile of the bile ducts or gallbladder. They are typically categorized into cholesterol, pigment, and mixed types, with cholesterol stones being the most prevalent[1]. Globally, approximately 6% of the population is affected by GSs, highlighting a significant public health concern. Despite a low mortality rate, over 20% of GS patients develop biliary symptoms or complications that require surgical intervention[1]. The complications include acute cholecystitis[2], GS pancreatitis[3], cholangitis due to common bile duct stones, GS-induced bowel obstruction[4], and even gallbladder cancer[5]. Over 50000 cholecystectomies are performed annually in the United Kingdom[6], and approximately 800000 in the United States, with an annual expenditure of nearly 6.5 billion dollars[7]. The substantial health and economic burden of GSs underscores the critical need for effective diagnosis, treatment, and prevention strategies.
GS formation has traditionally been attributed to factors such as supersaturated cholesterol, cholestasis, biliary infection, and excessive mucin secretion[8,9]. However, emerging evidence suggests an essential role of bile microbiota in the development and progression of GSs[10,11]. The bile microbiota, previously considered a healthy biliary system as essentially sterile[12], leading to the initial exclusion of microorganisms as contributors to the formation of GS, has gained attention due to advances in sequencing technologies such as 16S rRNA gene sequencing, which has revealed a diverse microbial community within the bile ducts. Molinero et al[10] in 2019 used 16S sequencing to describe the differences in biliary microflora between GS patients and healthy people by collecting bile from healthy liver donors. Interactions between bile microorganisms and host physiology may influence GS formation by altering bile acid composition[13] and cholesterol levels[14] or through direct interactions with gallbladder epithelial cells[15].
Evidence indicates that specific bacterial species or microbial communities in bile are associated with β-glucuronidase expression[16]. β-glucuronidase hydrolyzes bilirubin glucuronide into bilirubin and glucuronic acid, potentially leading to bilirubin calcium precipitation in the presence of calcium. Indeed, bacteria expressing β-glucuronidase are frequently identified in samples from patients with pigment GSs[17]. Additionally, Yu et al[15] have found that Helicobacter pylori cytotoxin-associated gene A promotes GS formation by increasing the permeability of gallbladder epithelial cells. Some lactic acid bacteria, including Lactobacillus acidophilus and Lactobacillus fermentum, can lower lipid, total cholesterol, and low-density lipoprotein (LDL) cholesterol levels in serum and liver[18]. Since cholesterol supersaturation is essential for GS formation, lowering cholesterol and lipids can be valuable preventive strategy.
However, research on the roles of specific bacteria in the formation of GSs in bile remains insufficient. With the growing recognition of the role of bile microbiome in various gastrointestinal and hepatobiliary diseases, elucidating the relationship between bile microbiota and GS formation are crucial for developing new preventive and therapeutic strategies. Our present study was designed to investigate the association between the bile microbiome and GS formation, pinpointing specific bacteria involved. Leveraging advanced sequencing technologies and comprehensive microbiome analyses, we explored how microbial diversity and composition related to GS development. Our findings are expected to enhance understanding of GS disease and highlight the importance of bile microbiota in hepatobiliary health.
Twenty-five patients diagnosed with GSs according to the European Association for the Study of the Liver guidelines[19] were assigned to the GS group and 15 patients diagnosed with non-stone benign gallbladder conditions such as gallbladder polyps and gallbladder adenomyomatosis were assigned to the GS-free (GSF) group. These conditions were selected as they represent common indications for cholecystectomy in the absence of GSs and typically involve minimal active inflammation compared to acute cholecystitis, providing a clinically relevant comparative group undergoing the same surgical procedure. The laparoscopic or open cholecystectomies were performed electively in the Department of General Surgery at Jinshan Hospital, Fudan University. This study was conducted in accordance with the ethical principles of the Helsinki Declaration (2013) and received approval from the Medical Ethics Committee of Jinshan Hospital, Fudan University. All participants provided informed consent. The medical records of the patients were comprehensively reviewed for documented history of prior biliary events (e.g., cholecystitis and pancreatitis), significant systemic infections within the past year, and chronic medical conditions (e.g., diabetes and metabolic syndrome). Detailed medication history, regarding agents known or suspected to impact the microbiome (including but not limited to antibiotics and probiotics), was extracted. These agents included proton pump inhibitors, metformin, statins, immunosuppressants, and chronic anti-inflammatory drugs. The three-month antibiotic/probiotic washout period prior to surgery was strictly applied based on documented prescriptions and patient recall during pre-operative assessment.
The inclusion criteria were: (1) Patients diagnosed with GSs by ultrasound, abdominal computed tomography (CT), or magnetic resonance cholangiopancreatography; and (2) Patients diagnosed with benign gallbladder diseases such as gallbladder polyps or gallbladder adenomyomatosis (but without GSs) by ultrasound, abdominal CT, or magnetic resonance cholangiopancreatography.
The exclusion criteria were: (1) Patients diagnosed with gallbladder cancer based on postoperative pathology; (2) Patients with a history of acute cholecystitis or acute exacerbation of chronic cholecystitis within the past three months; (3) Patients who experienced severe infection within the past three months; (4) Patients with gastrointestinal diseases or metabolic disorders such as type 2 diabetes, severe obesity, and hyperlipidemia; (5) Patients with severe diseases of vital organs including heart, liver, lungs, kidneys, and brain; and (6) Patients who had used antibiotics and probiotics within the past three months.
Fresh bile samples were collected intraoperatively from 27 patients, including 21 with GSs and six without GSs. After cholecystectomy, 10 mL of bile was aseptically aspirated from the gallbladder using a syringe and transferred into sterile containers. The samples were immediately flash-frozen in liquid nitrogen and stored at -80 °C for further processing, with proper labeling. They were then kept on dry ice for sequencing analysis. All samples were stored in their original cryotubes at -80 °C until further handling.
DNA extraction: Total microbial genomic DNA was extracted from bile samples using the PureLinkTM Microbiome DNA Purification Kit (A29790, Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The quality was assessed using 1% agarose gel electrophoresis.
DNA amplification by the polymerase chain reaction: Specific primers with barcodes were synthesized for targeted sequencing regions. To ensure the accuracy and reliability for subsequent data analysis, polymerase chain reaction (PCR) amplification was performed with low cycle numbers, and the consistency in the number of cycles per sample was maintained. Representative samples were randomly selected for preliminary experiments to ensure the vast majority of samples amplified the product of appropriate concentration in the lowest cycle number of appropriate concentration in the lowest cycle number. PCR was conducted using TransStart Fastpfu DNA Polymerase (AP221-02, TransGen, Beijing, China). All samples were processed according to the formal experimental conditions, with each sample being analyzed in triplicates. PCR products derived from the same sample were mixed and analyzed using 2% agarose gel electrophoresis. They were recovered using AxyPrep DNA Gel Extraction Kit (AP-GX-250, Axygen, Corning, NY, United States), eluted by Tris (hydroxymethyl) aminomethane hydrochloride, and detected by 2% agarose electrophoresis.
Fluorescence quantification: Based on the preliminary quantification results of electrophoresis, the PCR products were further detected and quantified using the QuantiFluorTM-ST Blue Fluorescence Quantification System (Promega, Madison, WI, United States). Subsequently, the corresponding proportions were mixed according to the sequencing volume requirements of each sample.
Library construction: The Illumina official connector sequences were added to the outer end of the target region by PCR. The PCR products were recovered by cutting the gel using a gel recovery kit, followed by Tris (hydroxymethyl) aminomethane hydrochloride, buffer elution and 2% agarose gel electrophoresis. Denaturation with sodium hydroxide yielded single-stranded DNA fragments. The reagents used for library construction was TruSeqTM DNA Sample Prep Kit (Illumina, San Diego, CA, United States).
Sequencing: One end of the DNA fragment was complementary to the primer base and immobilized on the chip. The DNA fragments were used as the template, and the base sequence fixed on the chip was used as the primer for PCR synthesis, and the target DNA fragments were synthesized on the chip. After denaturation and annealing, the other end of DNA fragments on the chip was randomly complementary to another nearby primer, also fixed, forming a “bridge”. DNA clusters were generated through PCR amplification. The DNA amplicons were linearized into single strands. The modified DNA polymerase and dNTPs with four fluorescent labels were added, synthesizing one base per cycle. The surface of the reaction plate was scanned with laser, and the nucleotide species aggregated in the first reaction of each template sequence were read. The “fluorescence group” and “termination group” were chemically cut, and the viscosity of the 3’ end was restored before continuing the polymerization of the second nucleotide. The fluorescence signal results collected in each round were counted to know the sequence of the template DNA fragment.
PE reads obtained from sequencing were first assembled based on overlap relationships, followed by sequence quality control and filtering. After the samples were distinguished, operational taxonomic units (OTU) clustering and taxonomic classification were conducted. Various diversity indices and sequencing depth were assessed based on the OTU clustering results. Additionally, principal coordinate analysis based on Bray-Curtis distance estimated the β-diversity of species composition. Statistical community structure analysis was performed at various taxonomic levels using classification information. Based on these analyses, a series of in-depth statistical and visual analyses, including multivariate analysis and difference significance test of community composition and phylogenetic information, were conducted.
The resulting data were used for species annotation and classification to generate the OTU tables using Uparse (version 11), RDP Classifier (version 2.13), and GreenGenes (version 135). Diversity analyses, including alpha diversity (e.g., Shannon index) and beta diversity (e.g., Bray-Curtis distance matrix), were performed using Qiime (version 1.9.1) and Mothur (version 1.30.2). Differential abundance analysis used statistical tools such as LEfSe to identify significant differences in microbial communities between the GS and GSF groups. Statistical analyses were performed using the R software package (version 3.3.1) and GraphPad Prism (version 9.5). The data are expressed as mean ± SD. Normality was assessed using the Shapiro-Wilk W-test. If the data met the assumptions of normality and homogeneity of variance, Student’s t-test was used; otherwise, the Mann-Whitney U-test was employed for group comparisons. In all cases, a
A total of 40 patients (25 in the GS group and 15 in the GSF group) were included in this study. The demographics of the participants are summarized in Table 1. No significant differences in age, gender distribution, or body mass index were observed between the two groups.
| Parameter | GS | GSF | P value |
| Frequency (n) | 25 | 15 | - |
| Gender | 0.327 | ||
| Male | 11 (44) | 9 (60) | |
| Female | 14 (56) | 6 (40) | |
| Age (years), mean ± SD | 53.480 ± 15.097 | 50.130 ± 11.661 | 0.466 |
| BMI (kg/m²), mean ± SD | 23.274 ± 2.247 | 25.015 ± 4.364 | 0.168 |
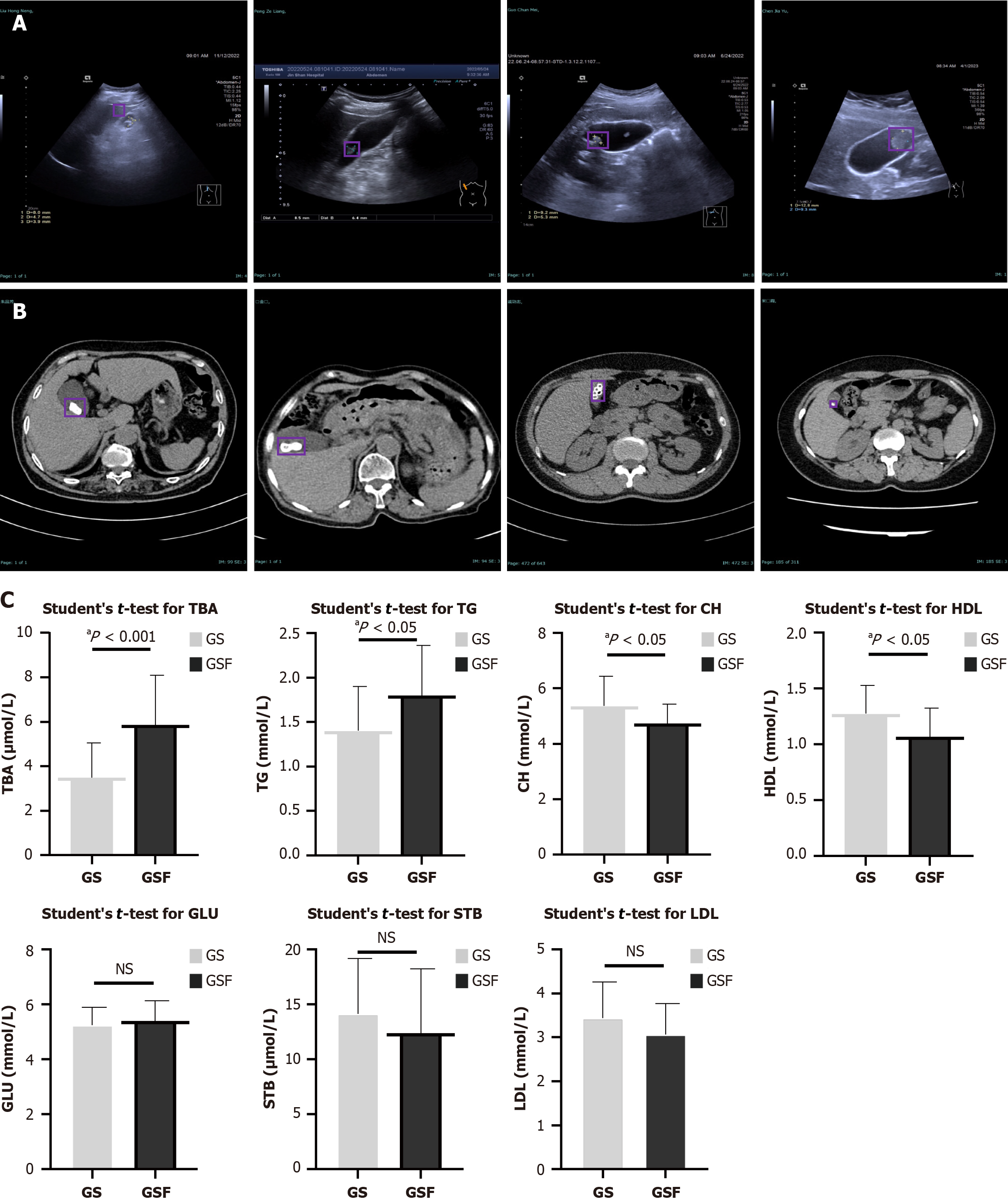
The diagnosis of GSs depends on abdominal ultrasound and CT. The typical ultrasound appearance of GSs is one or more fixed-shaped echogenic masses, echogenic spots, or arc-shaped strong echoes, often accompanied by posterior acoustic shadowing and capable of shifting with changes in patient positioning, as shown in Figure 1A. High-density shadows within the gallbladder on CT can suggest the presence of GSs (Figure 1B). A comparison of serum biochemical parameters revealed significant differences between the GS and GSF groups. As shown in Figure 1C, the GS group exhibited significantly lower levels of total bile acids (P < 0.001) and triglycerides (P < 0.05) compared to the GSF group. Conversely, cholesterol (P < 0.05) and high-density lipoprotein (P < 0.05) levels were significantly higher in the GS group. No significant differences were noted in glucose, serum total bilirubin, and LDL levels between the two groups (all P > 0.05). These laboratory examinations supported the possible mechanism of GS formation, such as cholesterol supersaturated and insufficient bile acid secretion.
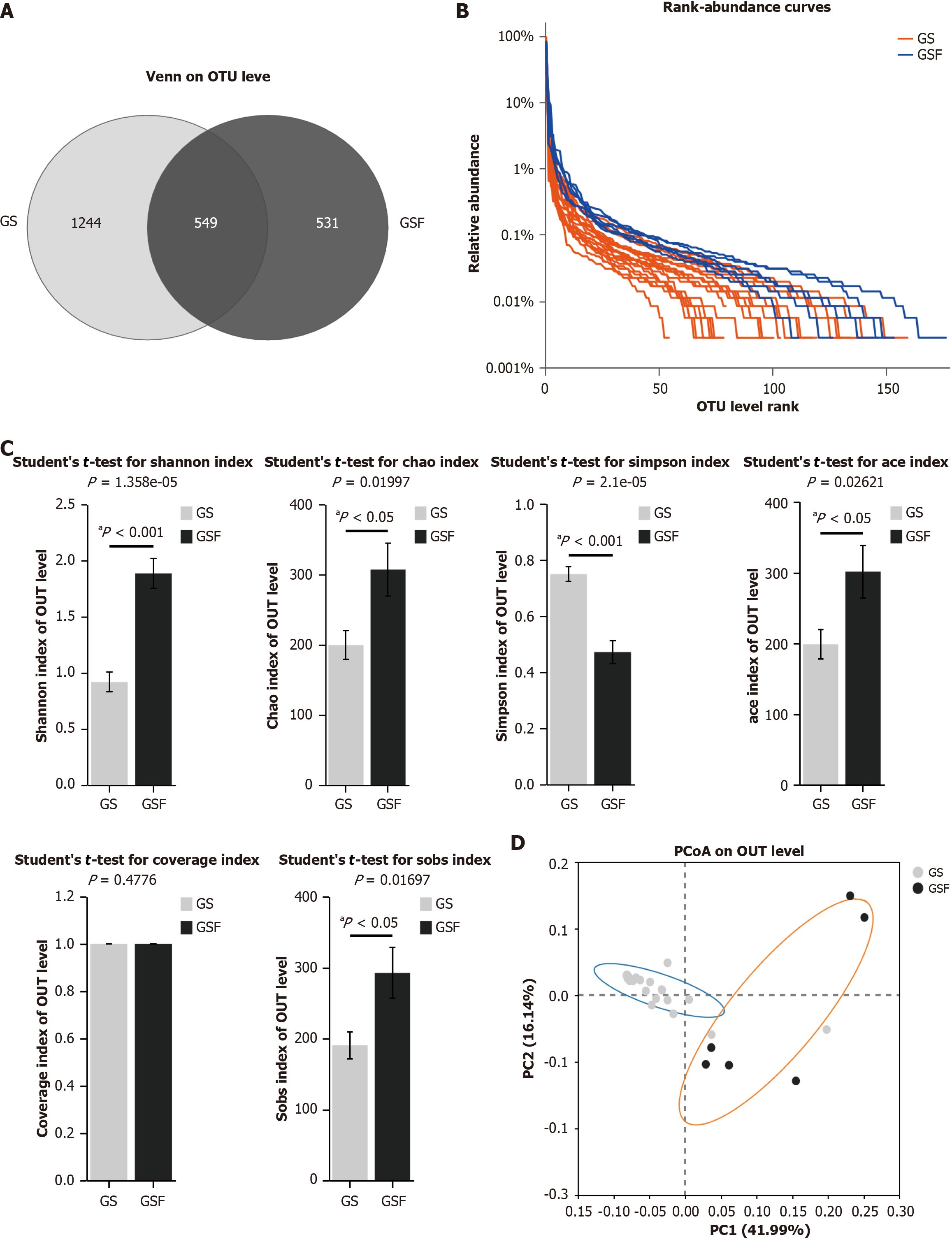
Results of 16S rRNA sequencing: The GSF group had 1080 OTUs, with 531 unique ones. In contrast, the GS group contained 1793 OTUs, with 1244 unique ones (Figure 2A). The Rank-Abundance curves reveal that the curve in GSF group declines more smoothly than in the GS group (Figure 2B), suggesting that GS group had a higher proportion of dominant microbial taxa and lower overall diversity.
Changes in bile microbiome diversity: Comparison of the intra-sample diversity between GS and GSF groups showed that the Shannon, Chao, Ace, and Sobs diversity index were significantly lower in the GS group than in the GSF group (Student’s t-test, all P < 0.05) (Figure 2C). In contrast, the Simpson index was significantly higher in the GS group than in the GSF group (Student’s t-test, P < 0.001). There was no significant difference in the Coverage index between the two groups. Overall, the bile microbiome diversity was markedly lower in the GS group than in the GSF group.
Differences in the bile microbiome between the two groups were evaluated using beta diversity. An unweighted Bray-Curtis distance matrix was generated to assess microbial community differences between the bile samples of the two groups. Principal coordinate analysis was employed to examine the similarity between samples. We observed that the microbiome communities from the bile of patients in the GS group clustered separately from those from the GSF group (Figure 2D).
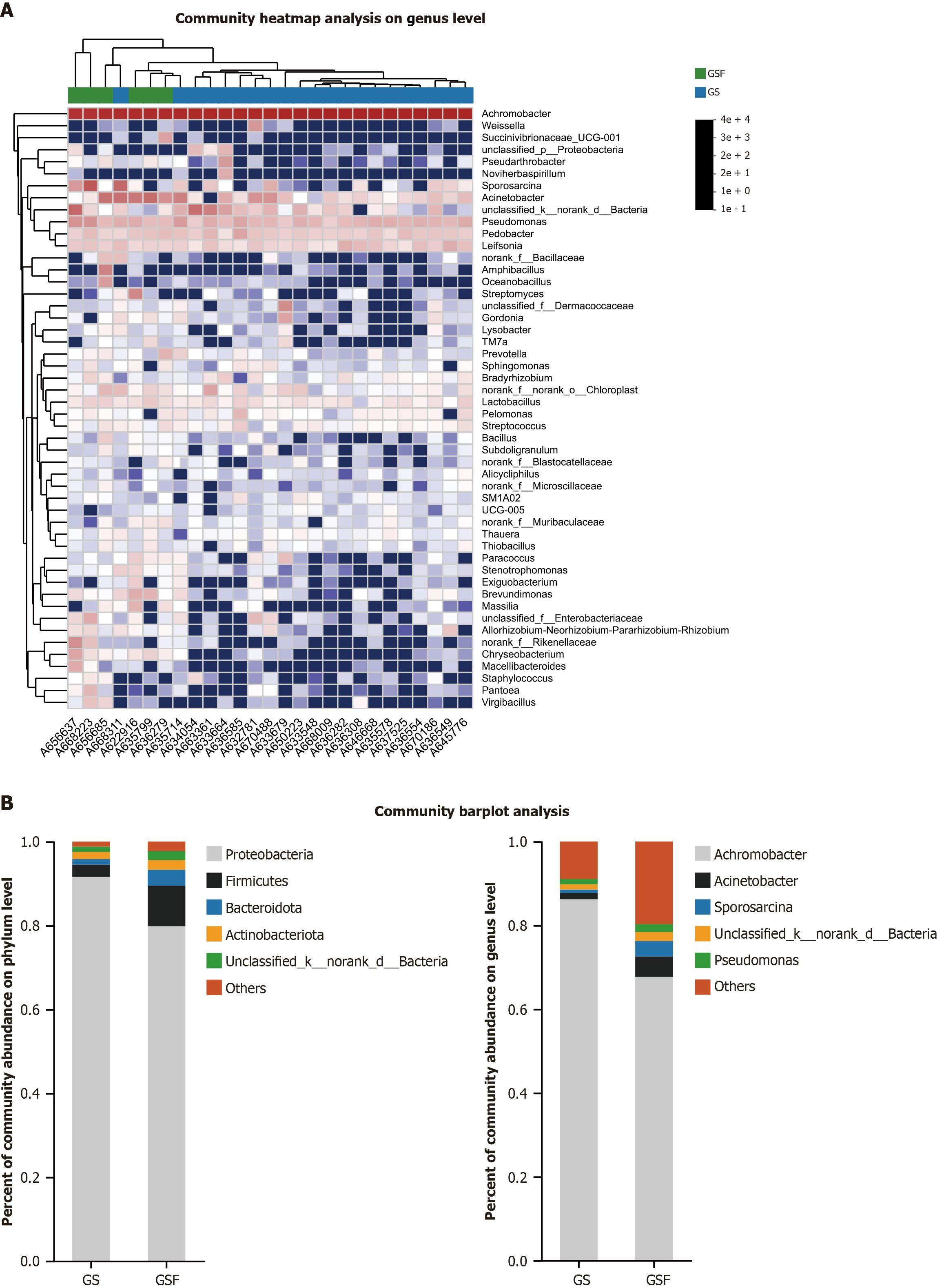
Composition and differences of bile microbiome: Taxonomic analysis identified several bacterial taxa that were differentially abundant between the GS and GSF groups. The heat map from the top 50 mean abundance data showed significant differences between the GS patient and GSF groups (Figure 3A). At the phylum level, the most abundant taxa in the bile microbiota of GS group were Proteobacteria (91.59%), followed by Firmicutes (2.90%) and Actinobacteria (1.70%). In contrast, the bile microbiota in the GSF group was predominantly composed of Proteobacteria (79.81%), Firmicutes (9.67%), and Bacteroidota (3.80%). At the genus level, Achromobacter (86.22%) was the most prevalent in the bile microbiota of GS group, followed by Acinetobacter (1.48%) and Pseudomonas (1.27%). In the bile microbiota of GSF group, Achromobacter was also predominant (67.68%), and other notable genera included Acinetobacter (4.86%) and Sporosarcina (3.71%) (Figure 3B).
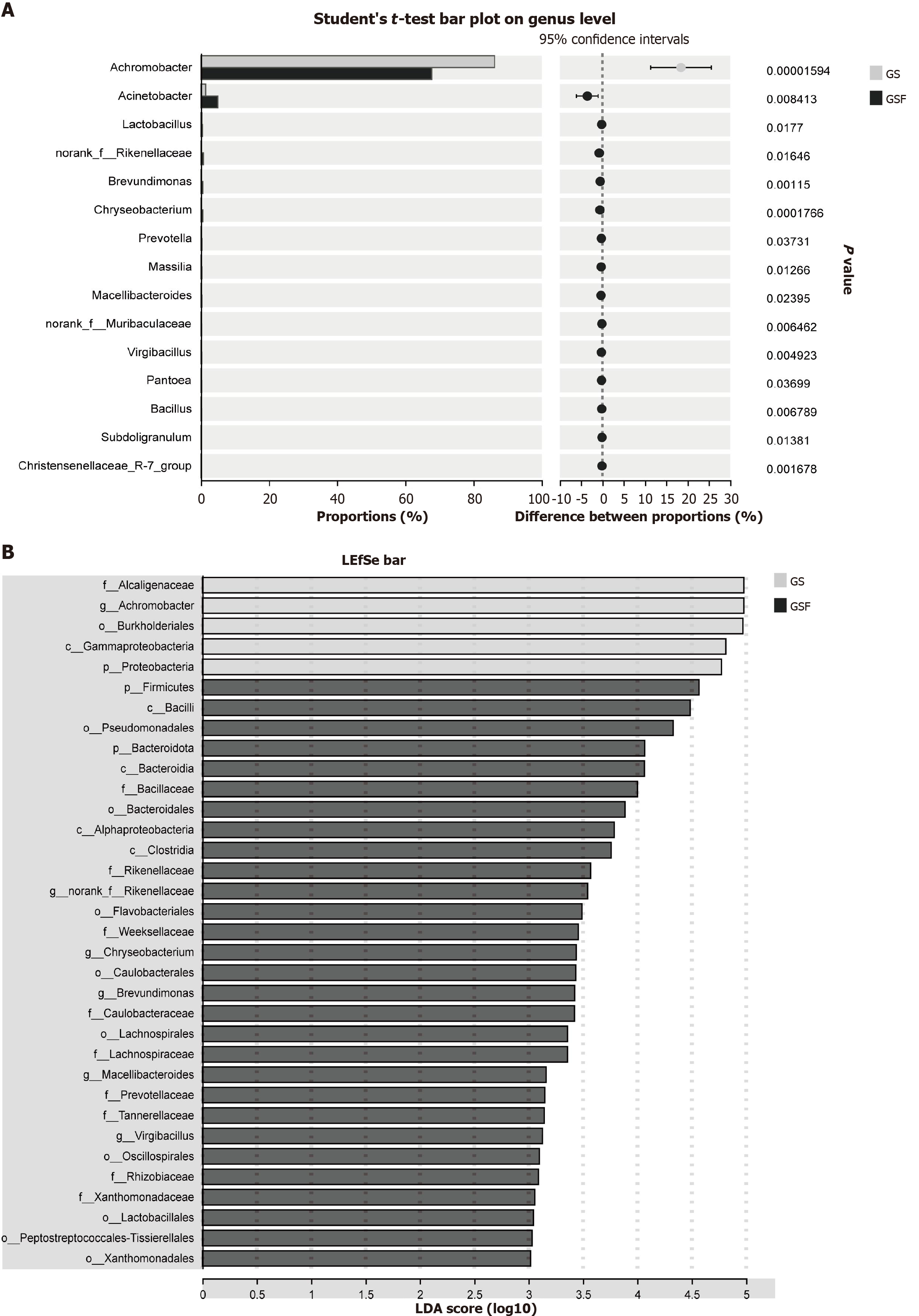
Differential analysis revealed a significantly higher abundance of Achromobacter in the bile of GS group, suggesting its potential as a biomarker for GSs. Conversely, genera such as Acinetobacter, Lactobacillus, and Prevotella were more abundant in the GSF group, implying that these genera might play beneficial roles and mitigate GS formation, as shown in Figure 4.
Extensive research has explored the relationship between the gastrointestinal microbiome and GSs[20,21], yet few studies have delved into biliary microbiology due to the challenges in obtaining bile samples. A series of studies have revealed the potential role of biliary microbiota in various diseases such as hepatobiliary tumors[22,23], primary sclerosing cholangitis[24], and giant common bile duct stones[25]. In our present study, bioinformatic analysis identified a distinct microbial community within the bile microbiome of GS patients. Overall, the bile microbiota diversity in the GS group was reduced compared to the GSF group. The bile microbiome was composed of four major phyla: Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidota. Additionally, the microbiome was characterized by a dominant genus, Achromobacter, and several other taxa with similar abundances, including Acinetobacter, Pseudomonas, and Sporosarcina. These findings aligned with several previous studies[10,11].
At the phylum level, the abundance of Proteobacteria was significantly higher in the GS group than in the GSF group, and the Firmicutes and Bacteroidetes were considerably lower than in the GSF group. The idea that an expansion of Proteobacteria is a microbial signature of epithelial dysfunction may explain these phenomena[26]. GS formation is also associated with epithelial dysfunction[27]. The effective absorption of fluid and nonpolar lipids from bile by the gallbladder epithelium is a normal physiological process that can prevent cholesterol supersaturation, and this process is often severely compromised in the presence of cholesterol GS disease or at least one of its precursors[28], which partly explains the high abundance of Proteobacteria in GS patients. At the genus level, we observed that the abundances of Acinetobacter, Lactobacillus, and Prevotella were lower in the GS group compared to the GSF group, implying possible beneficial roles of these genera in preventing GS formation. Lactobacillus is a widely recognized probiotic that improves human health and prevents various diseases[29]. Studies have reported that Lactobacillus acidophilus ATCC 43121 and Lactobacillus fermentum MF27 positively influence serum biochemical parameters in LDL-induced C57BL/6J mice after four weeks of treatment without affecting growth rates[14], which is believed to be mediated by reduced liver expression of β-Hydroxy β-methylglutaryl-CoA reductase. Furthermore, these characteristics may lead to decreased expression of gel-forming mucins (including MUC5AC and MUC5B) in the gallbladder. Both strains of lactic acid bacteria might prevent cholesterol GS formation in LDL-induced C57BL/6J mice[14,30]. Clinically, the regular intake of these lactic acid bacteria may help prevent cholesterol stone formation in the gallbladder. However, there are few reports on whether Acinetobacter and Prevotella affect the formation of GSs. Abdullah et al[31] isolated a variety of bacteria from GS cases, including the Gram-negative bacterium Acinetobacter. Cai et al[32] found Prevotella was rich in the diseased group, which was contrary to the results of our present study. Further experiments are needed to verify the specific roles of these two bacteria in the formation of GSs.
Recent studies suggest that alterations in the gut microbiota can affect bile acid composition and metabolism, which are essential for maintaining cholesterol solubility[19]. Thus, an imbalance in bile acid composition due to microbial dysbiosis may lead to cholesterol supersaturation and subsequent GS formation[19]. Our findings provide evidence supporting this mechanistic link. Specifically, total bile acids levels were significantly lower in the GS group than in the GSF group (P < 0.001). Conversely, total cholesterol (P < 0.05) and high-density lipoprotein (P < 0.05) levels were significantly elevated in the GS group.
Our study has several limitations that warrant consideration. Firstly, the retrospective design and modest sample size limit the statistical power and generalizability of our findings. Selection bias inherent in recruiting patients undergoing cholecystectomy may restrict applicability to the broader population with GSs, particularly asymptomatic cases or those managed conservatively. Furthermore, despite our efforts to capture and adjust for key confounders, the retrospective nature limits our ability to fully account for all potential influences on the bile microbiome, such as undocumented medication use, subtle prior infections, or long-term dietary patterns. Prospective studies with meticulously recorded longitudinal clinical data, extended washout periods for a broader range of medications where feasible, and standardized dietary assessments would provide a more robust control for confounding factors. Therefore, our results should be interpreted as preliminary associations specific to this surgical cohort, necessitating validation in larger, prospective studies encompassing diverse patient populations and disease stages. Secondly, we did not dig deeply into the mechanisms via which the biliary bacteria exert their roles, specifically in the composition ratio of bile acids and cholesterol. It is crucial to emphasize that the associations reported here are based on taxonomic profiles. The specific metabolic functions of the identified bile microbiota within the gallbladder environment and their direct mechanistic contributions (e.g., alterations in bile acid composition/deconjugation, cholesterol crystallization kinetics, or epithelial interactions) remain speculative based on the current data. Future research must move beyond taxonomy to investigate the functional metagenomics and metatranscriptomics of the bile microbiome in GS diseases. Furthermore, targeted in vitro studies (e.g., co-culture of representative bacterial strains with cholesterol crystals or gallbladder epithelial cells) and carefully designed in vivo models are essential to validate the hypothesized roles of specific bacteria (such as Achromobacter or Acinetobacter) and elucidate the precise molecular mechanisms underlying their potential contribution to cholesterol crystallization, inflammation, or stone nucleation/growth. Thirdly, laparoscopic/open cholecystectomy is an invasive procedure and it is not possible to obtain healthy bile by this method. A significant limitation is that our GSF control group had no GSs, but consisted of patients with other benign gallbladder conditions (gallbladder polyps, gallbladder adenomyomatosis). It is possible, though less likely than in inflammatory conditions, that these pathologies themselves could subtly influence the bile microbiota composition. While this represents the most feasible surgical control group available, we acknowledge that findings comparing GS patients to these non-stone controls may reflect differences associated with GSs and/or the specific underlying pathology of the control group. Therefore, attributing microbial differences solely to GS presence requires caution. Future studies exploring the bile microbiome in truly asymptomatic individuals (e.g., via rare intraoperative sampling during unrelated procedures, if ethically approved) would be highly valuable, though logistically difficult.
Understanding the characteristics of the bile microbiome is of increasing importance for the prevention and treatment of GSs. Several studies have explored the possibility of treating or preventing GSs by changing the microbial composition[14], such as utilizing antibiotic therapy or probiotic intervention to modulate the balance of bile microbiome. Future studies should further explore the exact role of microorganisms in the formation and development of GSs and develop effective strategies for improving patient outcomes by regulating microbial composition.
In summary, our study highlights significant differences in bile microbiota composition and diversity between GS and GSF patients. The higher abundance of proteobacteria Achromobacter in GS patients, coupled with reduced microbial diversity, suggests that alterations in bile microbiota may play a role in GS formation. Although these findings offered novel insights into bile microbiota dysbiosis in GS patients in our cohort, further validation in larger prospective studies is required to confirm generalizability and explore causality.
The authors acknowledge the support from the Department of General Surgery, Jinshan Hospital, Fudan University, and the Clinical Research Center Laboratory, Jinshan Hospital, Fudan University.
| 1. | Lammert F, Gurusamy K, Ko CW, Miquel JF, Méndez-Sánchez N, Portincasa P, van Erpecum KJ, van Laarhoven CJ, Wang DQ. Gallstones. Nat Rev Dis Primers. 2016;2:16024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 579] [Article Influence: 57.9] [Reference Citation Analysis (1)] |
| 2. | Gallaher JR, Charles A. Acute Cholecystitis: A Review. JAMA. 2022;327:965-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 226] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 3. | Wang GJ, Gao CF, Wei D, Wang C, Ding SQ. Acute pancreatitis: etiology and common pathogenesis. World J Gastroenterol. 2009;15:1427-1430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 178] [Cited by in RCA: 281] [Article Influence: 16.5] [Reference Citation Analysis (5)] |
| 4. | Alemi F, Seiser N, Ayloo S. Gallstone Disease: Cholecystitis, Mirizzi Syndrome, Bouveret Syndrome, Gallstone Ileus. Surg Clin North Am. 2019;99:231-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 5. | Ryu S, Chang Y, Yun KE, Jung HS, Shin JH, Shin H. Gallstones and the Risk of Gallbladder Cancer Mortality: A Cohort Study. Am J Gastroenterol. 2016;111:1476-1487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 6. | Beckingham IJ. ABC of diseases of liver, pancreas, and biliary system. Gallstone disease. BMJ. 2001;322:91-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 80] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 7. | Shaffer EA. Epidemiology and risk factors for gallstone disease: has the paradigm changed in the 21st century? Curr Gastroenterol Rep. 2005;7:132-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 323] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 8. | Wang HH, Portincasa P, Afdhal NH, Wang DQ. Lith genes and genetic analysis of cholesterol gallstone formation. Gastroenterol Clin North Am. 2010;39:185-207, vii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 9. | Di Ciaula A, Wang DQ, Portincasa P. An update on the pathogenesis of cholesterol gallstone disease. Curr Opin Gastroenterol. 2018;34:71-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 176] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 10. | Molinero N, Ruiz L, Milani C, Gutiérrez-Díaz I, Sánchez B, Mangifesta M, Segura J, Cambero I, Campelo AB, García-Bernardo CM, Cabrera A, Rodríguez JI, González S, Rodríguez JM, Ventura M, Delgado S, Margolles A. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome. 2019;7:100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 11. | Hu J, Tang J, Zhang X, Yang K, Zhong A, Yang Q, Liu Y, Li Y, Zhang T. Landscape in the gallbladder mycobiome and bacteriome of patients undergoing cholelithiasis with chronic cholecystitis. Front Microbiol. 2023;14:1131694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Csendes A, Burdiles P, Maluenda F, Diaz JC, Csendes P, Mitru N. Simultaneous bacteriologic assessment of bile from gallbladder and common bile duct in control subjects and patients with gallstones and common duct stones. Arch Surg. 1996;131:389-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 90] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 13. | Petrov VA, Fernández-Peralbo MA, Derks R, Knyazeva EM, Merzlikin NV, Sazonov AE, Mayboroda OA, Saltykova IV. Biliary Microbiota and Bile Acid Composition in Cholelithiasis. Biomed Res Int. 2020;2020:1242364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 14. | Oh JK, Kim YR, Lee B, Choi YM, Kim SH. Prevention of Cholesterol Gallstone Formation by Lactobacillus acidophilus ATCC 43121 and Lactobacillus fermentum MF27 in Lithogenic Diet-Induced Mice. Food Sci Anim Resour. 2021;41:343-352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 15. | Yu J, He Y, Yao W, Liu T, Liu X, Zheng Y, Hao C, Xue D. Helicobacter pylori CagA Promotes the Formation of Gallstones by Increasing the Permeability of Gallbladder Epithelial Cells. Helicobacter. 2024;29:e13100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 16. | Maki T. Pathogenesis of calcium bilirubinate gallstone: role of E. coli, beta-glucuronidase and coagulation by inorganic ions, polyelectrolytes and agitation. Ann Surg. 1966;164:90-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 328] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 17. | Kaufman HS, Magnuson TH, Lillemoe KD, Frasca P, Pitt HA. The role of bacteria in gallbladder and common duct stone formation. Ann Surg. 1989;209:584-91; discussion 591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 92] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 18. | Kerlikowsky F, Müller M, Greupner T, Amend L, Strowig T, Hahn A. Distinct Microbial Taxa Are Associated with LDL-Cholesterol Reduction after 12 Weeks of Lactobacillus plantarum Intake in Mild Hypercholesterolemia: Results of a Randomized Controlled Study. Probiotics Antimicrob Proteins. 2025;17:1086-1095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 19. | European Association for the Study of the Liver (EASL). EASL Clinical Practice Guidelines on the prevention, diagnosis and treatment of gallstones. J Hepatol. 2016;65:146-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 444] [Cited by in RCA: 372] [Article Influence: 37.2] [Reference Citation Analysis (2)] |
| 20. | Hu H, Shao W, Liu Q, Liu N, Wang Q, Xu J, Zhang X, Weng Z, Lu Q, Jiao L, Chen C, Sun H, Jiang Z, Zhang X, Gu A. Gut microbiota promotes cholesterol gallstone formation by modulating bile acid composition and biliary cholesterol secretion. Nat Commun. 2022;13:252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 174] [Article Influence: 43.5] [Reference Citation Analysis (3)] |
| 21. | Liu X, Qi X, Han R, Mao T, Tian Z. Gut microbiota causally affects cholelithiasis: a two-sample Mendelian randomization study. Front Cell Infect Microbiol. 2023;13:1253447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 22. | Song X, Wang X, Hu Y, Li H, Ren T, Li Y, Liu L, Li L, Li X, Wang Z, Huang W, Bao R, Zhang Y, Li M, Wang X, Liu F, Gu J, Zheng L, Wu W, Liu Y. A metagenomic study of biliary microbiome change along the cholecystitis-carcinoma sequence. Clin Transl Med. 2020;10:e97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 23. | Di Carlo P, Serra N, D'Arpa F, Agrusa A, Gulotta G, Fasciana T, Rodolico V, Giammanco A, Sergi C. The microbiota of the bilio-pancreatic system: a cohort, STROBE-compliant study. Infect Drug Resist. 2019;12:1513-1527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 24. | Pereira P, Aho V, Arola J, Boyd S, Jokelainen K, Paulin L, Auvinen P, Färkkilä M. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS One. 2017;12:e0182924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 25. | Dai C, Xu C, Zheng L, Wang M, Fan Z, Ye J, Su D. Characteristics and metabolic potential of biliary microbiota in patients with giant common bile duct stones. Front Cell Infect Microbiol. 2023;13:1259761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 26. | Litvak Y, Byndloss MX, Tsolis RM, Bäumler AJ. Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol. 2017;39:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 473] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 27. | Hu FL, Chen HT, Guo FF, Yang M, Jiang X, Yu JH, Zhang FM, Xu GQ. Biliary microbiota and mucin 4 impact the calcification of cholesterol gallstones. Hepatobiliary Pancreat Dis Int. 2021;20:61-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 28. | Ginanni Corradini S, Yamashita G, Nuutinen H, Chernosky A, Williams C, Hays L, Shiffman ML, Walsh RM, Svanvik J, Della Guardia P, Capocaccia L, Holzbach RT. Human gallbladder mucosal function: effects on intraluminal fluid and lipid composition in health and disease. Dig Dis Sci. 1998;43:335-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Maftei NM, Raileanu CR, Balta AA, Ambrose L, Boev M, Marin DB, Lisa EL. The Potential Impact of Probiotics on Human Health: An Update on Their Health-Promoting Properties. Microorganisms. 2024;12:234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 149] [Article Influence: 74.5] [Reference Citation Analysis (0)] |
| 30. | Ye X, Huang D, Dong Z, Wang X, Ning M, Xia J, Shen S, Wu S, Shi Y, Wang J, Wan X. FXR Signaling-Mediated Bile Acid Metabolism Is Critical for Alleviation of Cholesterol Gallstones by Lactobacillus Strains. Microbiol Spectr. 2022;10:e0051822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 31. | Abdullah BH, Jassam SA, Hadi WA, Hameed B. Gallbladder stone formation in Iraqi patients is associated with bacterial infection and HLA class II-DRB1 antigens. Indian J Pathol Microbiol. 2020;63:570-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 32. | Cai X, Peng Y, Gong Y, Huang X, Liu L, Chen Y, Du J, Dai Z, Qian Y, Xu L. Variations of bile bacterial community alongside gallstone disease progression and key taxa involved in poor outcomes after endoscopic surgery. Eur J Med Res. 2023;28:313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/