Copyright
©The Author(s) 2025.
World J Gastrointest Surg. Jan 27, 2025; 17(1): 97148
Published online Jan 27, 2025. doi: 10.4240/wjgs.v17.i1.97148
Published online Jan 27, 2025. doi: 10.4240/wjgs.v17.i1.97148
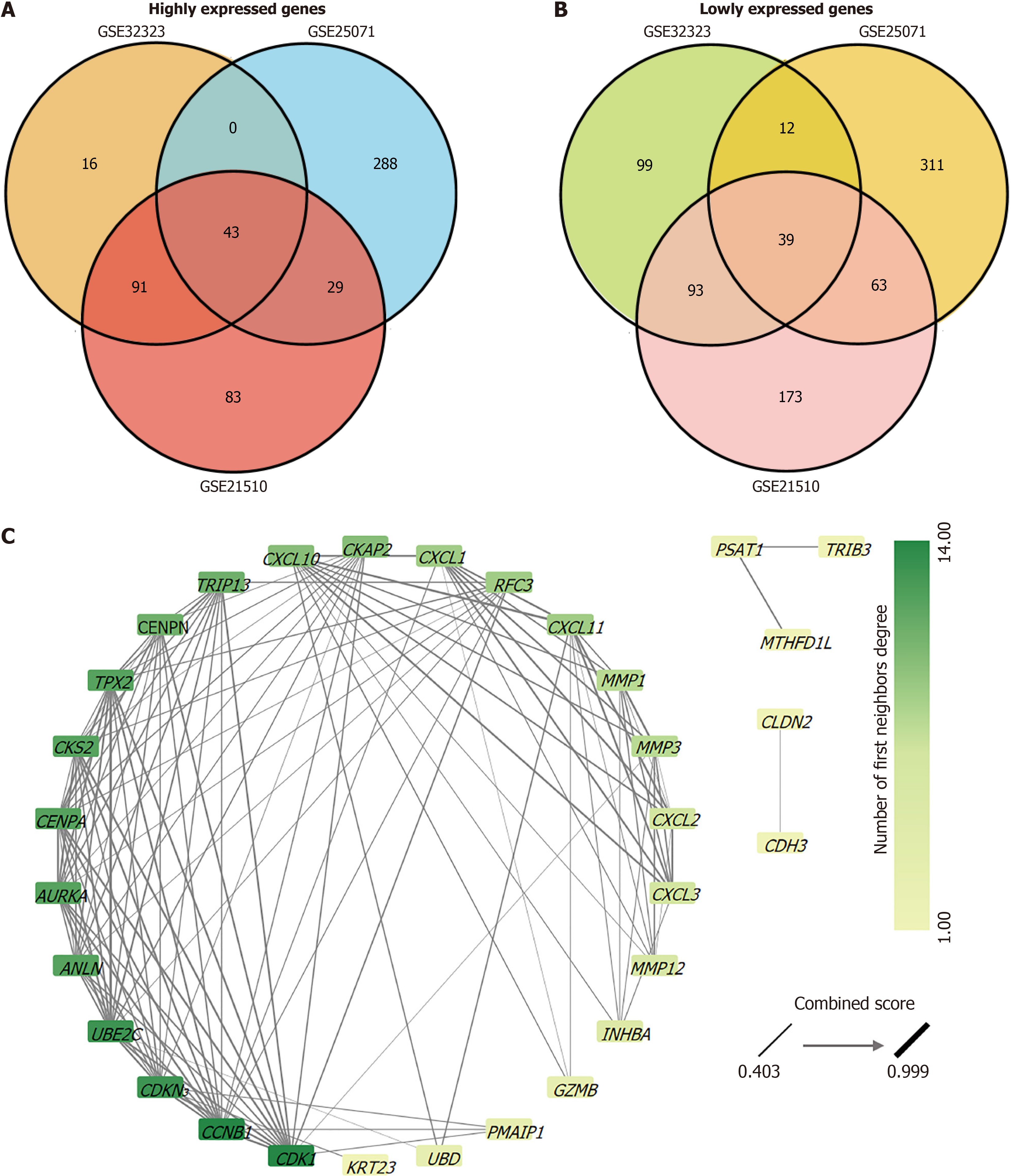
Figure 1 Protein-protein interaction network of highly expressed genes in colorectal cancer tissues.
A: A Venn diagram showed the number of relative highly expressed genes from the GSE32323, GSE25071, and GSE21510 datasets. These gene expressions were increased in colorectal cancer (CRC) tissues compared to normal tissues and an intersection of 43 genes was identified across all three datasets; B: A Venn diagram showed the number of genes with relatively low expression from GSE32323, GSE25071, and GSE21510. These gene expressions were decreased in CRC tissues compared to normal tissues and an intersection of 39 genes was identified across all three datasets. All genes were screened for |Log2 (fold change)|> 2 and adjusted P value < 0.05; C: The topology of the protein-protein interaction network was analyzed by Cytoscape and the network was established based on 43 genes from an intersection of relative overexpressed genes from the GSE32323, GSE25071, and GSE21510 datasets. The greater the adjacent node count, the darker the green; the thicker the edges, the stronger the interaction force.
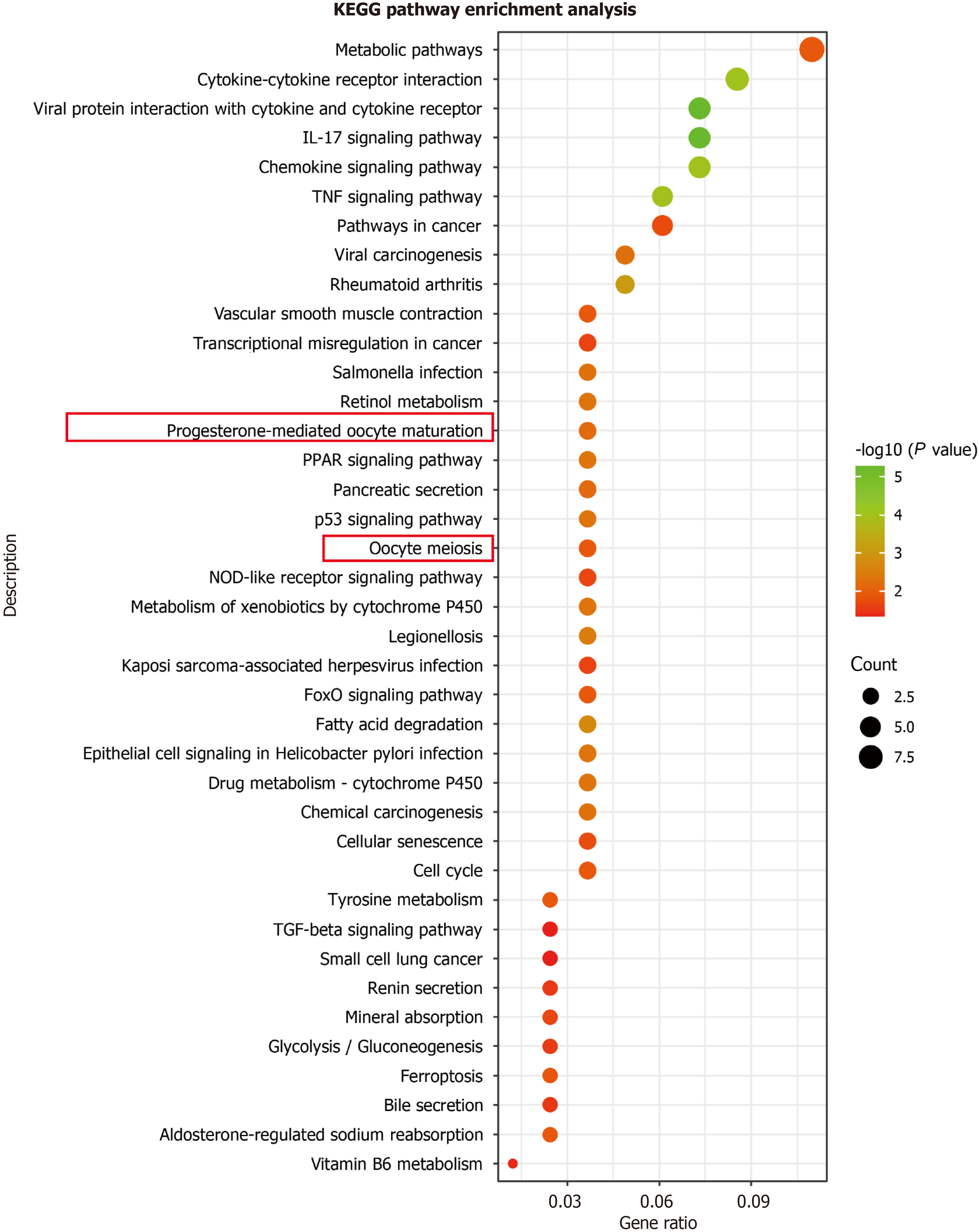
Figure 2 Bubble plot of Kyoto encyclopedia of genes and genomes pathway enrichment analysis results.
All pathways had a corrected P value < 0.05. The KOBAS 3.0 (http://bioinfo.org/kobas/genelist/) was used for Kyoto encyclopedia of genes and genomes pathway enrichment analysis of 43 genes with relatively high expression and 39 genes with relatively low expression. The order of the pathways was based on -log10 (P value). The red rectangle marks the pathways in which Aurora A participated. KEGG: Kyoto encyclopedia of genes and genomes; IL: Interleukin; TNF: Tumor necrosis factor; TGF: Transforming growth factor; PPAR: Peroxisome proliferators-activated receptor.
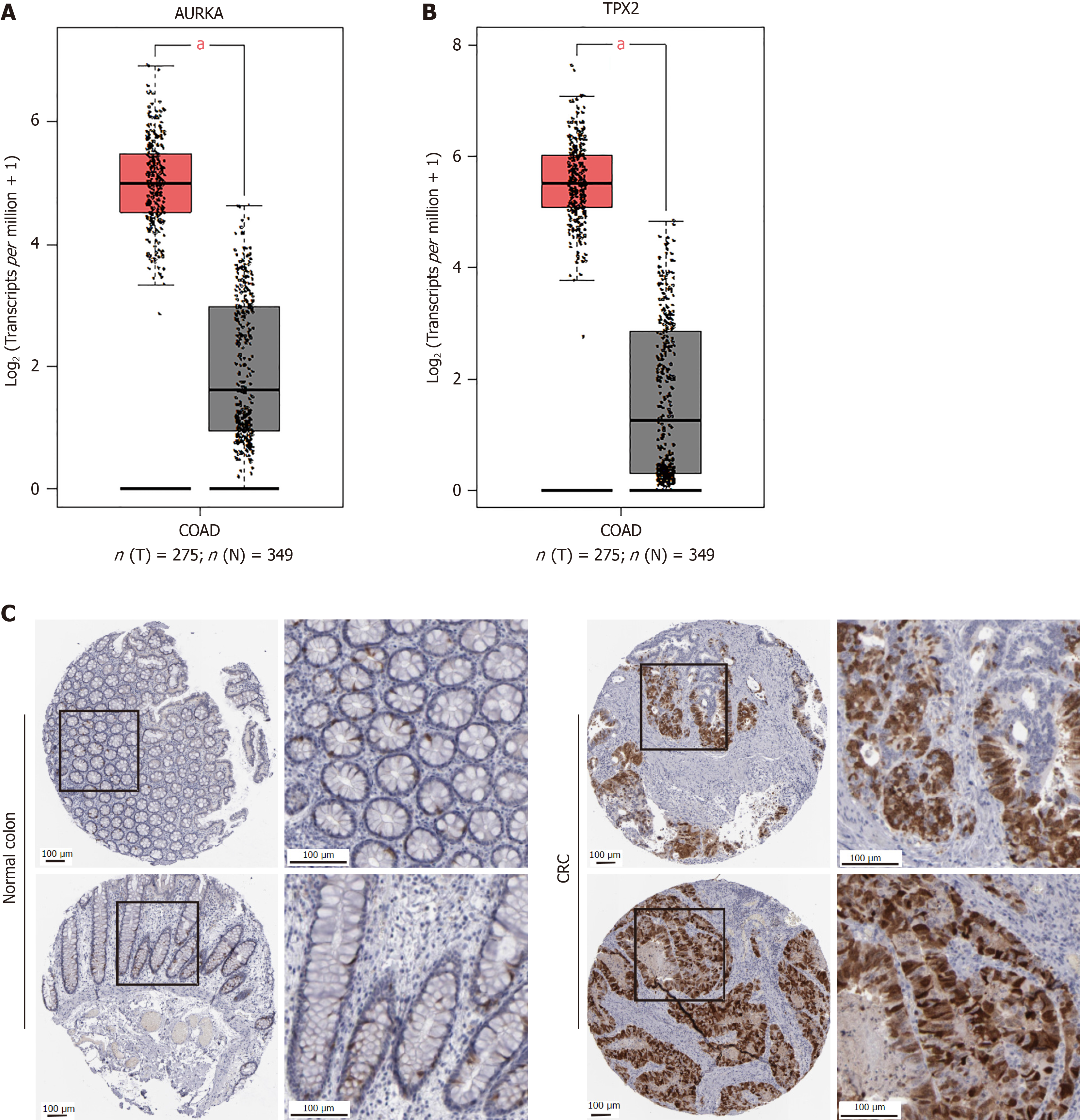
Figure 3 Aurora A and targeting protein for Xklp2 are high expressed in human colonic adenocarcinoma tissue.
A: The Aurora kinase A (AURKA) expression in colonic adenocarcinoma (COAD) tissue was increased compared to normal tissues; B: The targeting protein for Xklp2 expression in COAD tissue was increased compared to normal tissues. The expression level analysis results are from gene expression profiling interactive analysis; C: Expression of the AURKA protein in human COAD tissue sample or normal colon. The normal colon tissue photos from (https://www.proteinatlas.org/ENSG00000087586-AURKA/tissue/colonimg); the COAD tissue photos from the human protein atlas (https://www.proteinatlas.org/ENSG00000087586-AURKA/pathology/colorectal+cancerimg). aP < 0.05. AURKA: Aurora kinase A; TPX2: Targeting protein for Xklp2; COAD: Colonic adenocarcinoma; CRC: Colorectal cancer; T: Tumors; N: Normal.
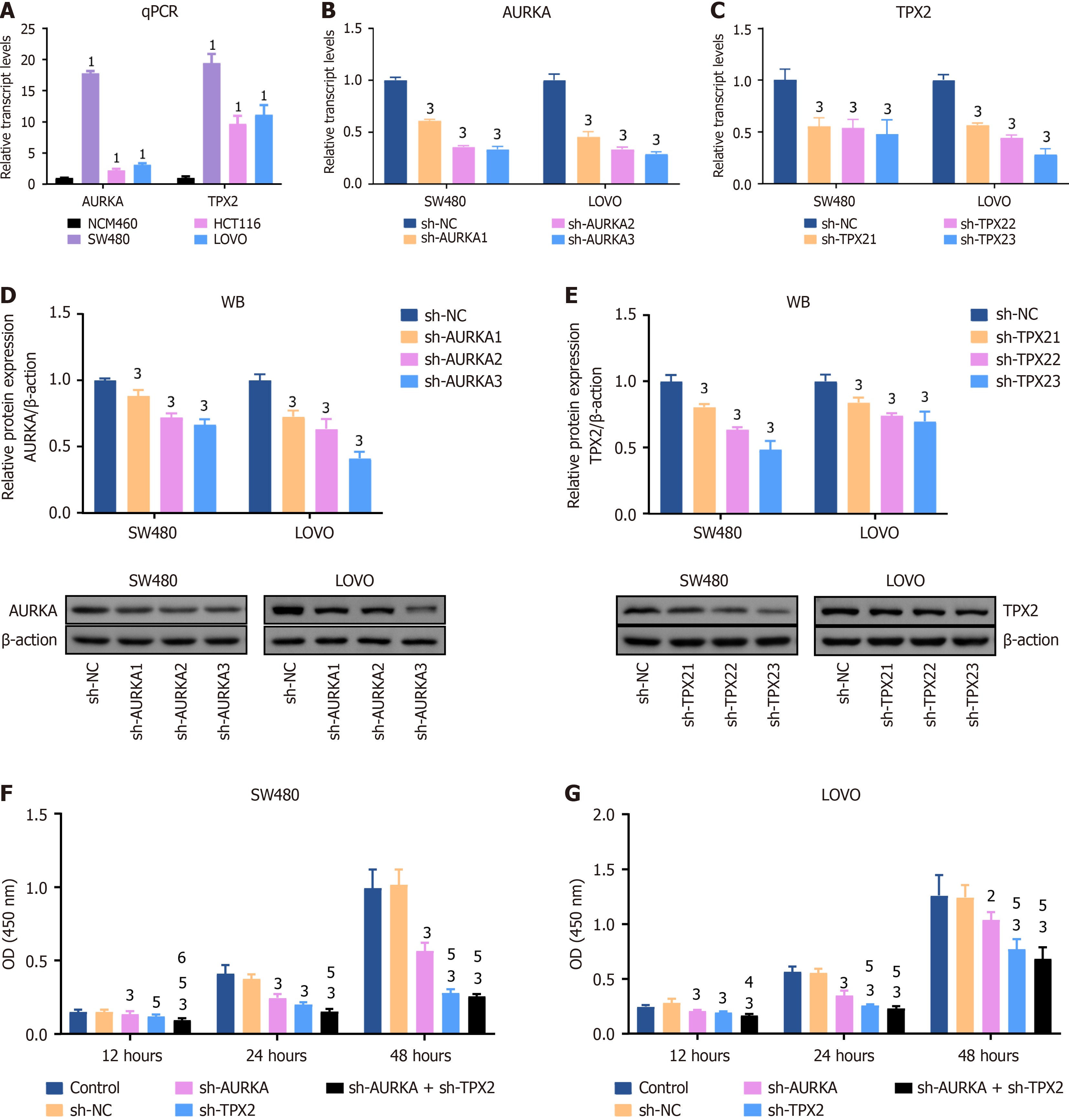
Figure 4 Knockdowns of Aurora A and targeting protein for Xklp2 inhibited viability of colorectal cancer cell.
A: Relative expression of Aurora kinase A (AURKA) and targeting protein for Xklp2 (TPX2) in NCM460, SW480, HCT116, and LOVO cells; B: After short hairpin RNA of AURKA (sh-AURKA) 1, 2, and 3 plasmids transfection, AURKA mRNA expression was decreased in SW480 and LOVO cells; C: TPX2 mRNA expression was decreased in SW480 and LOVO cells with transfected sh-TPX21, 2, and 3 plasmids. All gene transcript levels were measured by quantitative real time-polymerase chain reaction; D: AURKA; E: TPX2 protein expression levels were decreased in SW480 and LOVO cells with sh-AURKA/TPX21, 2, and 3 plasmids transfection. Western blotting was used to detect relative protein expression. Cell counting kit-8 assay was used to detect cell viability in F: SW480; G: LOVO cells with AURKA and TPX2 knockdown. After 12-, 24-, or 48-hour treatment with transfection, cell viability was inhibited in SW480 and LOVO cells. 1P < 0.01 vs NCM460 group. 2P < 0.05 vs negative control of short hairpin RNA (sh-NC) group. 3P < 0.01 vs sh-NC group. 4P < 0.05 vs short hairpin RNA of AURKA (sh-AURKA) group. 5P < 0.01 vs sh-AURKA group. 6P < 0.01 vs short hairpin-TPX2 group. qPCR: Quantitative real time-polymerase chain reaction; AURKA: Aurora kinase A; TPX2: Targeting protein for Xklp2; sh-NC: Negative control of short hairpin RNA; sh-AURKA: Short hairpin RNA of Aurora kinase A; sh-TPX2: Short hairpin RNA of targeting protein for Xklp2; OD: Optical density; WB: Western blotting.
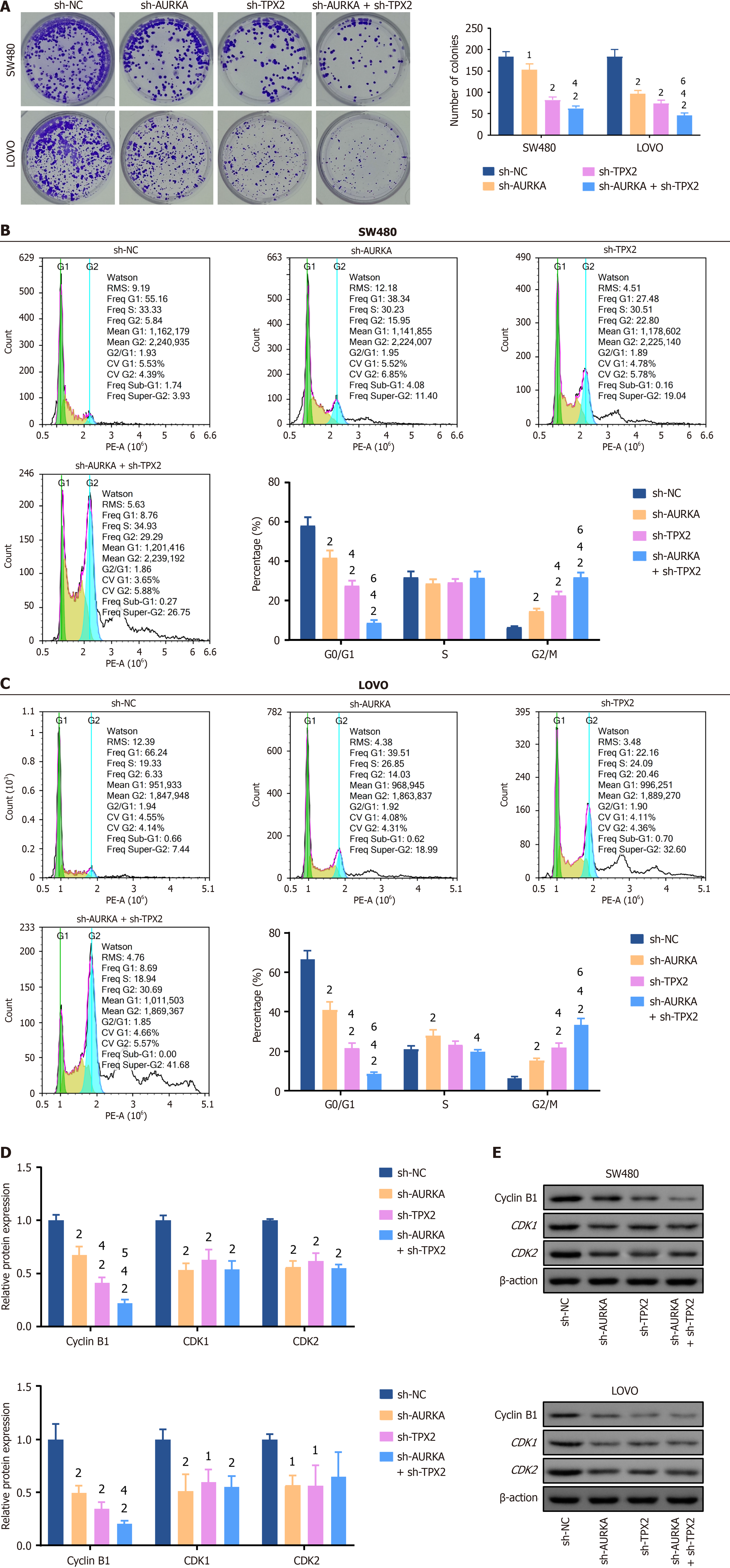
Figure 5 Knockdowns of Aurora A and targeting protein for Xklp2 inhibited colorectal cancer cell proliferation ability and decreased cell percentage in the G0/G1 phase.
A: The cell cloning assay was used to determine cell proliferation ability. Knockdown or co-knockdown of Aurora kinase A and targeting protein for Xklp2 (TPX2) inhibited the number of cell colonies; B: Cell cycle in SW480 detected by flow cytometry; C: Cell cycle in LOVO cells was detected by flow cytometry. Knockdown or co-knockdown AURKA and TPX2 decreased cell percentage in the G0/G1 phase; D: Western blotting was used to detect the expression levels of cell cycle-related biomarkers. Cyclin B1, CDK1, and CDK2 expressions were decreased in SW480 and LOVO cells with AURKA and TPX2 co-knockdown; E: The representative bands of western blotting. 1P < 0.05 vs negative control of short hairpin RNA (sh-NC) group. 2P < 0.01 vs sh-NC group. 3P < 0.05 vs short hairpin RNA of AURKA (sh-AURKA) group. 4P < 0.01 vs sh-AURKA group. 5P < 0.05 vs short hairpin-TPX2 (sh-TPX2) group. 6P < 0.01 vs sh-TPX2 group. AURKA: Aurora kinase A; TPX2: Targeting protein for Xklp2; sh-NC: Negative control of short hairpin RNA; sh-AURKA: Short hairpin RNA of Aurora kinase A; sh-TPX2: Short hairpin RNA of targeting protein for Xklp2.
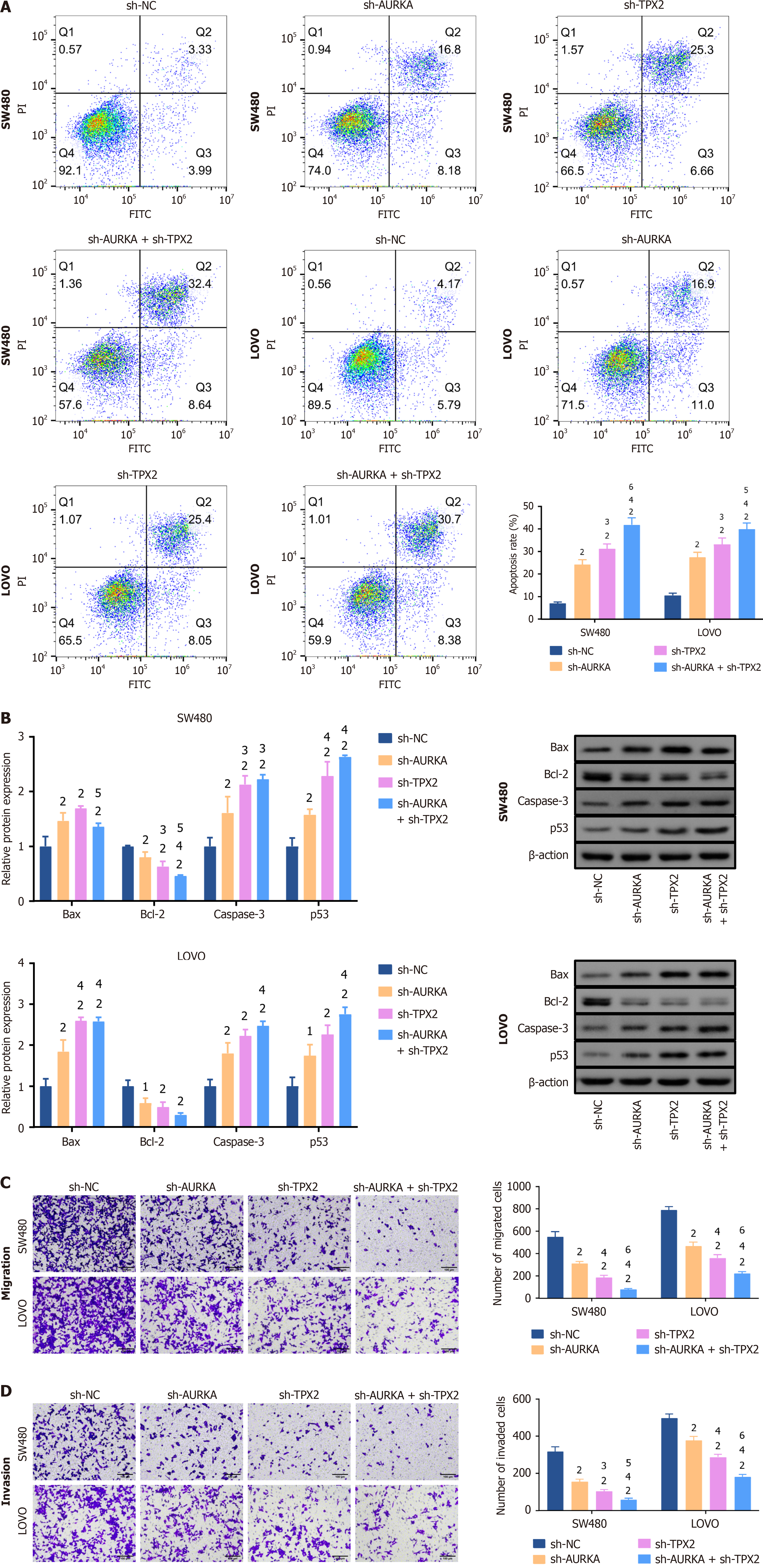
Figure 6 Knockdowns of Aurora A and targeting protein for Xklp2 promoted colorectal cancer cell apoptosis and inhibited cell migration and invasion.
A: Cell apoptosis in SW480 and LOVO cells was determined by flow cytometry. Apoptosis cells were increased in Aurora kinase A (AURKA)/targeting protein for Xklp2 (TPX2) knockdown or co-knockdown cells; B: Western blotting was used to detect the expression levels of apoptosis-related biomarkers. B-cell lymphoma-2 (Bcl-2)-associated X protein, caspase 3, and tumor protein P53 expression levels were increased in SW480 and LOVO cells with AURKA/TPX2 knockdown or co-knockdown, while Bcl-2 was decreased; C: The number of migrated cells; D: Invaded cells was decreased in SW480 and LOVO cells with AURKA/TPX2 knockdown or co-knockdown. Transwell assay was used to detect cell migration and invasion abilities. 1P < 0.05 vs negative control of short hairpin RNA (sh-NC) group. 2P < 0.01 vs sh-NC group. 3P < 0.05 vs short hairpin RNA of AURKA (sh-AURKA) group. 4P < 0.01 vs sh-AURKA group. 5P < 0.05 vs short hairpin-TPX2 (sh-TPX2) group. 6P < 0.01 vs sh-TPX2 group. AURKA: Aurora kinase A; TPX2: Targeting protein for Xklp2; sh-NC: Negative control of short hairpin RNA; sh-AURKA: Short hairpin RNA of Aurora kinase A; sh-TPX2: Short hairpin RNA of targeting protein for Xklp2; Bcl-2: B-cell lymphoma-2; Bax: B-cell lymphoma-2 associated X protein; p53: Tumor protein P53; FITC: Fluorescein isothiocyanate; PI: Propidium iodide.
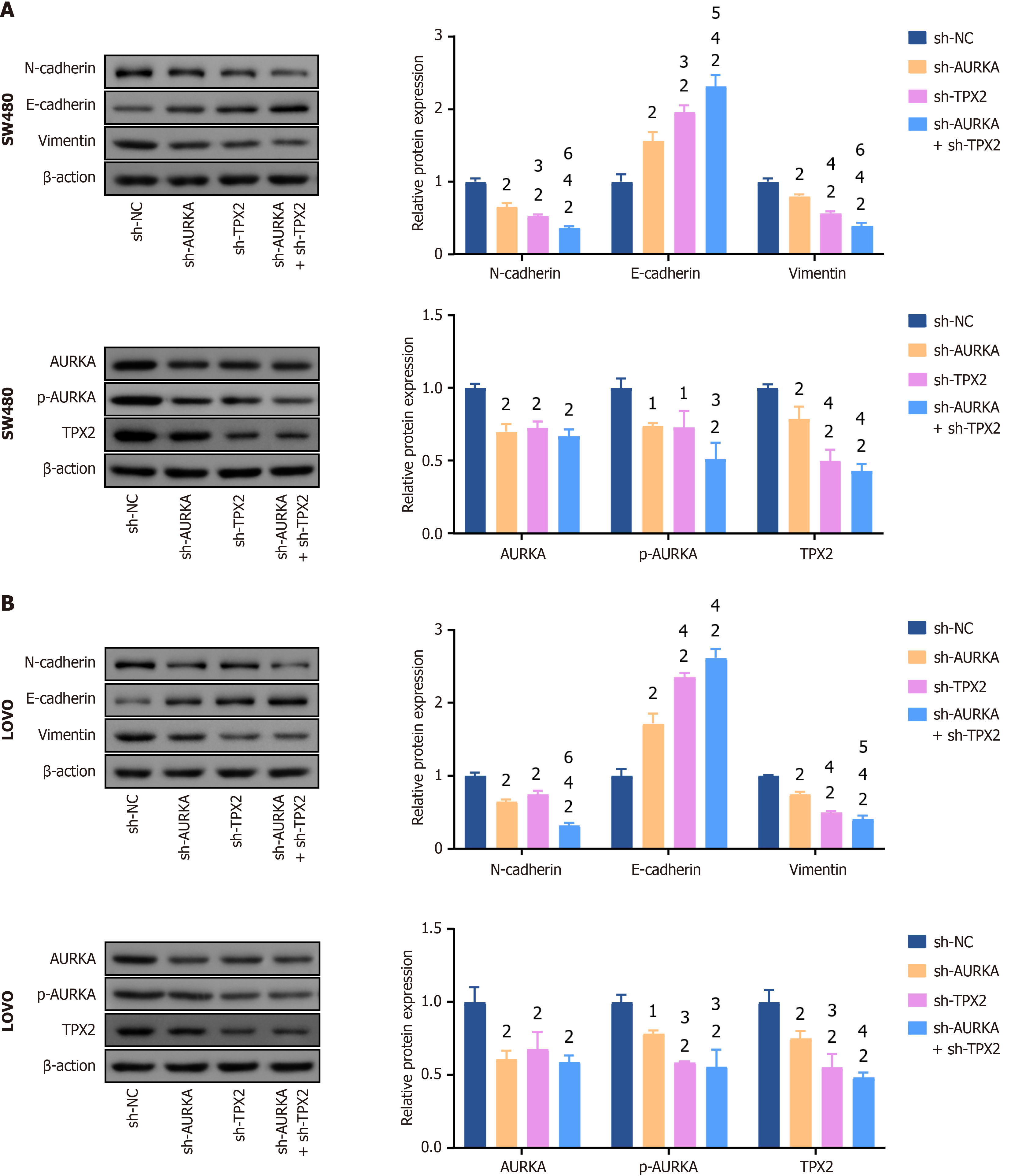
Figure 7 Knockdowns of Aurora A and targeting protein for Xklp2 inhibited epithelial-mesenchymal transition and knockdowns of Aurora A/targeting protein for Xklp2 pathways.
A: Knockdowns of Aurora kinase A (AURKA)/targeting protein for Xklp2 (TPX2) or co-knockdown in SW480; B: AURKA/TPX2 knockdown or co-knockdown LOVO cells decreased N-cadherin, vimentin, AURKA, phosphorylated AURKA, and TPX2 expression, and increased E-cadherin expression. Epithelial-mesenchymal transition biomarkers include N-cadherin, E-cadherin, and vimentin. Western blotting was used to detect relative protein expression. 1P < 0.05 vs negative control of short hairpin RNA (sh-NC) group. 2P < 0.01 vs sh-NC group. 3P < 0.05 vs short hairpin RNA of AURKA (sh-AURKA) group. 4P < 0.01 vs sh-AURKA group. 5P < 0.05 vs short hairpin-TPX2 (sh-TPX2) group. 6P < 0.01 vs sh-TPX2 group. AURKA: Aurora kinase A; TPX2: Targeting protein for Xklp2; sh-NC: Negative control of short hairpin RNA; sh-AURKA: Short hairpin RNA of Aurora kinase A; sh-TPX2: Short hairpin RNA of targeting protein for Xklp2; p-AURKA: Phosphorylated Aurora kinase A.
- Citation: Sheng GX, Zhang YJ, Shang T. Synergistic inhibition of colorectal cancer progression by silencing Aurora A and the targeting protein for Xklp2. World J Gastrointest Surg 2025; 17(1): 97148
- URL: https://www.wjgnet.com/1948-9366/full/v17/i1/97148.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v17.i1.97148