修回日期: 2024-04-06
接受日期: 2024-05-22
在线出版日期: 2024-06-28
长链非编码RNA小核仁RNA宿主基因16(long non-coding RNA small nucleolar RNA host gene 16, LncRNA SNHG16), 是小核仁RNA宿主基因家族的成员, 主要位于细胞质中. 研究表明SNHG16在前列腺癌、乳腺癌、膀胱癌、胃癌 、结肠癌等多种恶性肿瘤中异常高表达. 通过多种途径调控肿瘤细胞增殖、侵袭、迁移和凋亡等. 本文主要总结了SNHG16在消化系统恶性肿瘤中的作用, 包括SNHG16的异常表达、分子功能、调控网络以及相关临床特征, 为肿瘤的早期诊断和早期治疗提供新思路.
核心提要: 消化系统恶性肿瘤发病隐匿, 早期诊断困难, 预后差, 寻找高特异性和敏感性的肿瘤标志物对于肿瘤的诊治至关重要, 近年研究发现长链非编码小核仁RNA宿主基因16(long non-coding RNA small nucleolar RNA host gene 16, LncRNA SNHG16)是参与肿瘤发生发展的关键因素, 本文就SNHG16在消化系统恶性肿瘤中的研究进展做一综述.
引文著录: 叶惠, 李明月, 施瑞华. 长链非编码RNA SNHG16在消化系统肿瘤中作用机制研究进展. 世界华人消化杂志 2024; 32(6): 405-411
Revised: April 6, 2024
Accepted: May 22, 2024
Published online: June 28, 2024
The long non-coding RNA small nucleolar RNA host gene 16 (SNHG16), a member of the small nucleolar RNA host gene family, is mainly located in the cytoplasm. Studies have showed that SNHG16 is abnormally highly expressed in prostate cancer, breast cancer, bladder cancer, stomach cancer, colon cancer, and other malignant tumors. It influ-ences the proliferation, invasion, migration, and apoptosis of tumor cells through various mechanisms. In this paper, we summarize the role of SNHG16 in digestive system malignant tumors, including its abnormal expression, molecular function, regulatory network, and related clinical features, with an aim to provide new ideas for early diagnosis and early treatment of tumors.
- Citation: Ye H, Li MY, Shi RH. Advances in understanding of mechanism of long non-coding RNA SNHG16 in digestive system tumors. Shijie Huaren Xiaohua Zazhi 2024; 32(6): 405-411
- URL: https://www.wjgnet.com/1009-3079/full/v32/i6/405.htm
- DOI: https://dx.doi.org/10.11569/wcjd.v32.i6.405
癌症作为全球性重大公共卫生问题, 一直是世界范围内主要的死亡原因之一, 也是临床研究的重点[1]. 癌症的发生与基因突变、表观遗传改变、染色体易位、缺失和扩增密切相关[2]. 长链非编码RNA(long non-coding RNA, lncRNA)是一类长度大于200个核苷酸的线性非编码RNA, 可以从外显子、内含子、基因间区或5/3非翻译区转录, 并折叠成复杂的二级结构, 已便于与DNA、RNA和蛋白质的相互作用[3], 最终影响肿瘤细胞的增殖、侵袭、凋亡、转移与血管生成等. 因此, lncRNA可作为致癌或抑癌基因, 在人类肿瘤的发生发展中起着至关重要的作用.
长链非编码小核仁RNA宿主基因16(long non-coding RNA small nucleolar RNA host gene 16, LncRNA SNHG16)是小核仁RNA宿主基因家族的成员, 主要位于细胞质中, 最早发现于神经母细胞瘤中[4]. 随后的研究表明SNHG16在前列腺癌、乳腺癌、膀胱癌、胃癌、结肠癌等多种肿瘤中发挥重要作用[5]. 本文将对SNHG16在消化系统肿瘤中的作用机制及潜在应用展开综述, 以期为消化系统肿瘤的临床诊治提供新的思路和方向.
胃癌(gastric cancer, GC)是世界上最常见的恶性肿瘤之一, 也是癌症相关死亡的第四大原因[6]. 胃癌的发生与年龄、遗传、环境、饮食、饮酒、幽门螺旋杆菌感染等因素有关. 研究GC的潜在致病机制, 对于GC的早诊早治意义重大.
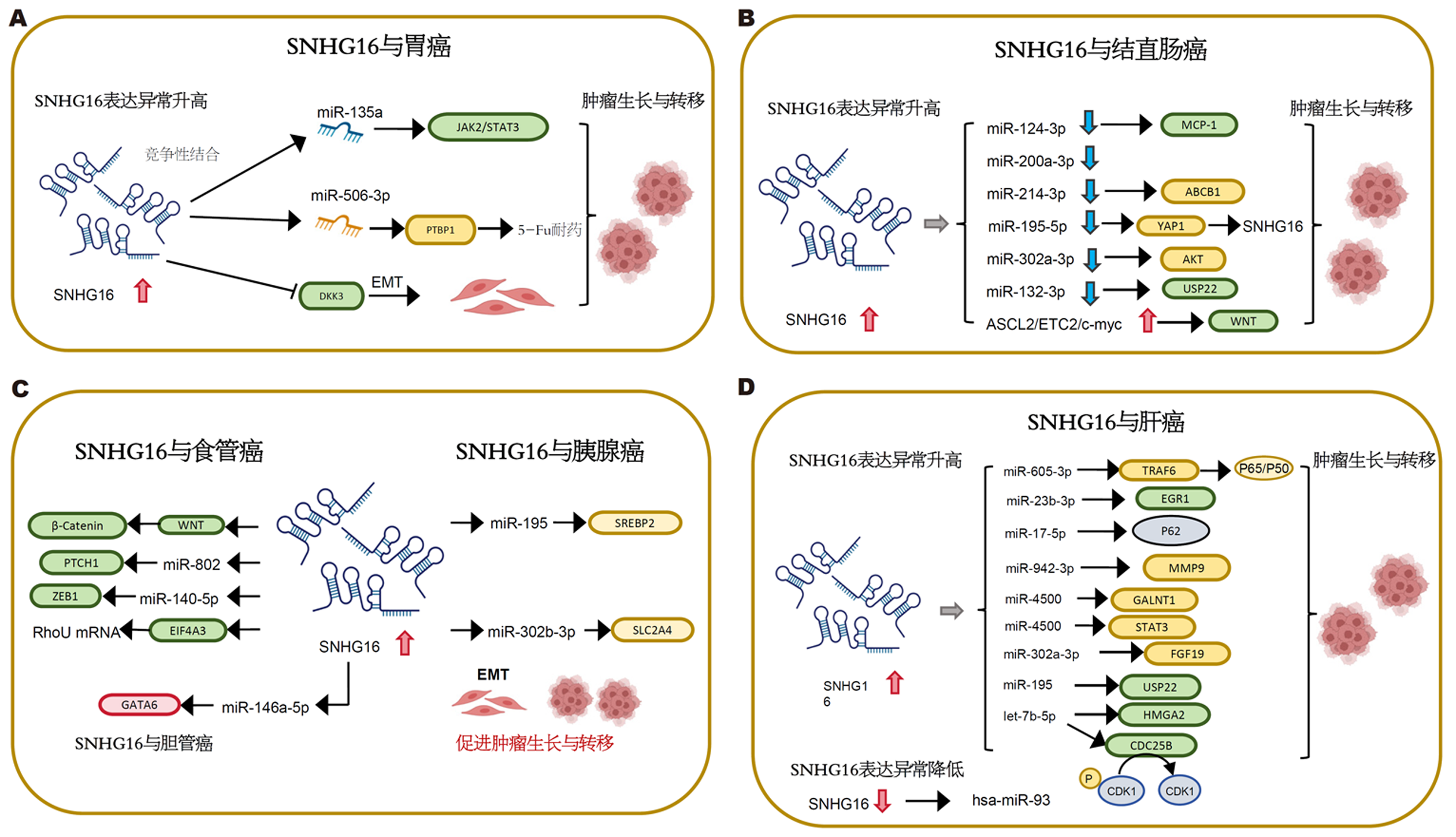
Lian等[7]最早发现SNHG16在胃癌组织中表达升高, 并通过影响细胞的增殖、侵袭、迁徙、凋亡, 发挥促癌作用. 随后, Wang等[8]的研究表明高水平的SNHG16与胃癌患者的肿瘤大小、TNM(Tumor node metastasis)分期呈正相关, 与生存率呈负相关. SNHG16作为内源性RNA, 通过与JAK2竞争性结合miR-135a, 启动JAK2/STAT3信号通路, 促进细胞增殖、集落形成和细胞侵袭. 此外, SNHG16通过miR-506-3p-PTBP1-葡萄糖代谢轴, 介导胃癌细胞的5-Fu耐药分子机制, 可能为5-Fu耐药的胃癌治疗提供新的治疗策略[9]. 胃癌患者预后与是否发生肿瘤转移密切相关, 恶性肿瘤细胞通过上皮间质转化(epithelial-mesenchymal transition, EMT)获得迁徙和侵袭能力, SNHG16通过下调GC细胞中WNT信号通路抑制剂3促进EMT过程, 因此SNHG16可能被作为预测GC患者转移的生物标志物[10]. 在GC细胞中, 抑制SNHG16表达, 显著下调c-Myc的表达, 影响p27/cyclin D1/CDK6、p53/cyclin E1和cyclin A2/CDK2复合物的形成, 导致细胞停滞在G1期, 加快细胞凋亡[11], 这些证据提示SNHG16也可能成为GC治疗的新靶点(图1A).
结直肠癌(colorectal cancer, CRC)是全球第三大最常见的恶性肿瘤, 也是导致癌症相关死亡的第二大原因[12]. 结直肠癌的发生涉及遗传因素、慢性炎症刺激和免疫功能障碍, 目前临床上对结直肠癌的治疗主要有手术、放疗和化疗, 深入研究结直肠癌发生发展的具体机制, 对寻找结直肠癌治疗特异性靶基因, 开发新的免疫治疗靶点, 以有效延长患者的生存率至关重要.
早在2016年, Christensen等[13]发现SNHG16在结直肠癌组织中较正常组织明显上调, SNHG16的表达受Wnt通路调控, 与Wnt通路调控的转录因子ASCL2、ETC2、c-myc的表达呈正相关, 从而影响肿瘤细胞的活力、凋亡和迁移. 随后, Li等[14]发现SNHG16通过靶向miR-200a-3p, 促进结直肠癌细胞的生长和侵袭. 然而, SNHG16通过具体何种机制影响结肠癌的发展还尚未知晓. Ke等[15]通过StarBase数据库预测, 确定miR-302a-3p是SNHG16的靶标, 通过miRanda数据库预测确定Akt是miR-302a-3p的靶标, 首次发现SNHG16通过靶向miR-302a-3p/AKT轴调控结肠癌细胞的增殖. 当SNHG16与miRNA的相互作用在结直肠癌中被证实, 更多miRNA被相继研究, 后续研究发现SNHG16同样可通过SNHG16/miR-132-3p/USP22通路[16]、SNHG16/miR-214-3p/ABCB1轴影响直肠癌细胞的生长、凋亡等[17]. 这些研究都表明, SNHG16在结直肠癌中起致癌作用. 此外, SNHG16也可通过miR-124-3p/MCP-1轴促进结直肠癌细胞迁移和EMT[18]. 肝脏是结直肠癌远处转移最常见的部位, SNHG16的表达与结直肠癌患者TNM分期、远处转移、预后不良相关. 一方面, SNHG16通过"海绵化"miR-195-5p, 保护YAP1免受抑制, 通过YAP1促进CRC增殖、迁移、侵袭以及肝脏转移; 另一方面, YAP1可通过形成转录复合物, 结合SNHG16启动子位点, 调控其转录; 形成SNHG16-YAP1/TEAD1正反馈回路[19], 上调SNHG16的表达. 综上所述, SNHG16可能是CRC的潜在治疗靶点(图1B).
食管癌是世界上第八大最常见的癌症类型, 也是导致癌症死亡的第六大原因. 其发生的危险因素包括巴雷特食管、胃食管反流、肥胖、吸烟、饮酒、热饮和亚硝胺的摄入等[20]. 食管癌死亡率高, 诊断预后差, 深入了解食管癌的致病机制, 提高预防和治疗策略, 对患者的早诊早治、生存改善至关重要.
有文献报道[21], SNHG16在食管癌组织和细胞系中表达上调, SNHG16的表达水平与肿瘤分期、淋巴结转移密切相关. SNHG16通过WNT/β-catenin途径, 影响食管癌细胞的增殖; 此外, SNHG16可作为食管癌患者总生存率的独立预测因子. RNA-RNA的相互作用在食管癌中也同样被多次研究, SNHG16作为食管癌中致癌lncRNA, 通过"海绵化"miR-140-5p, 调节靶基因ZEB1, 促进细胞的增殖、迁移和EMT[22]; 通过"海绵化"miR-802, 上调PTCH1, 激活Hedgehog通路, 促进食管癌细胞的自我更新[23]; 通过结合RNA结合蛋白-真核生物翻译起始因子(EIF4A3)并与之形成复合物, 联合调控Ras同源家族成员U mRNA的稳定性, 发挥促癌作用[24].
胰腺癌是一种高度侵袭性的恶性肿瘤, 有致命风险, 主要见于男性和老年人. 近年来, 胰腺癌患者年轻化, 由于疾病早期缺乏特异性症状, 严重影响患者预后, 确定早期诊断方案是提高胰腺癌的诊出和生存率的重要途径[25].
多项研究表明[26,27], SNHG16参与胰腺癌的发生发展. SNHG16在胰腺癌组织和细胞系中表达明显升高, 与肿瘤TNM分期、远处转移和不良预后密切相关. lncRNA最常通过"海绵化"miRNA, 发挥促癌或抑癌作用. SNHG16通过负向调节miR-302b-3p的表达, 直接靶向SLC2A4促进肿瘤进展[26]; 通过海绵吸附miR-195调节SREBP2的表达, 促进脂肪生成, 加速胰腺癌的进程[27]. 肿瘤耐药是肿瘤治疗的一大挑战, 最新的一项研究发现, SNHG16可通过EZH2介导的表观遗传修饰, 结合Smad4启动子下调其表达, 诱导胰腺癌的化疗耐药[28]. LncRNA与miRNA的相互作用以及参与转录水平调控, 为胰腺癌的诊断和治疗提供了新的见解.
胆管癌(cholangiocarcinoma, CCA)是人类常见的恶性肿瘤, 近年来发病率迅速上升, CCA可分为肝内胆管癌(intrahepatic cholangiocarcinoma, ICC)、肝门周围胆管癌(perihilar cholangiocarcinoma, pCCA)和远端胆管癌三种亚型. 目前手术是CCA最有效的治疗方法. 由于缺乏早期有效和敏感的诊断方法, 许多患者错过了治疗的最佳时间窗, 因此, 迫切需要寻找新的治疗方法来改善CCA患者的预后, 延长患者的生存时间.
Wu等[29]通过TCGA数据库中的信息, 分析CCA中SNHG16的表达. 与正常组织相比, SNHG16在癌组织中表达明显上调, 与CCA细胞系中的结果一致, 过表达SNHG16显著促进CCA细胞增殖, 抑制凋亡. 机制研究提示: SNHG16作为ceRNA, 通过直接靶向miR-146a-5p/GATA6轴在CCA中实现促瘤作用, 这可能为CCA的诊断和治疗提供新的见解, 打开了CCA后续研究的新思路. SNHG16在食管癌、胰腺癌、胆管癌中的作用机制见(图1C).
肝细胞癌(hepatocellular carcinoma, HCC)是最常见的原发性肝癌类型, HCC是导致癌症相关死亡的第二大原因[30]. 由于其高复发、易远处转移, 导致患者总生存期较差, 一直备受关注. 因此, 揭示HCC发生、进展、转移等全面而复杂的分子机制具有关键意义.
Guo等[31]通过逆转录PCR技术检测到肝癌组织和细胞系中SNHG16的表达增高, 其表达水平与肿瘤大小、甲胎蛋白水平、门静脉肿瘤血栓形成、转移相关. SNHG16高表达促进HCC细胞增殖、迁移和侵袭, 抑制细胞凋亡, 与患者预后不良相关, 可作为HCC的独立预后因素[32], 在机制上, SNHG16作为内源性RNA, 在HCC中同样参与多条通路调控肿瘤发展. SNHG16通过激活ECM受体相互作用途径促进HCC细胞恶性行为[33]. 通过作为ceRNA调控miR-186, 促进HCC细胞增殖、迁移和侵袭[34]; 通过结合miR-195, 起到"海绵化"作用, 导致其表达下调[35]; 通过海绵miR-4500和靶向STAT3表达促进HCC进展[36]; 通过SNHG16/miR-302a-3p/FGF19轴促进肝癌细胞的增殖[37]; 通过竞争性结合miR -17-5p, 抑制p62的表达, 促进HCC的[32]; 通过SNHG16/miR-605-3p/TRAF6/NF-κB通路, 调控HCC细胞的转移效应, 高表达的SNHG16能够活化NF-κB促进肿瘤转移, 反过来, 活化的NF-κB作用于SNHG16, 介导其上调, 形成正反馈回路[38], 这可能是HCC中SNHG16高表达的原因之一; SNHG16还可通过"海绵化"miR-23b-3p调控EGR1的表达, 调节Hep3B细胞的自噬;促进索拉非尼耐药[39]; 也可通过海绵化miR-140-5p诱导肝癌细胞索拉非尼的耐药[40]. SNHG16一方面通过直接作用于let-7b-5p/CDC25B/cdk1轴促进HCC细胞G2/M周期转变; 一方面通过let-7b-5p/HMGA2轴诱导细胞转移和EMT进展[41]; 另外, 外泌体被认为是组织微环境和循环系统中细胞间信号传递的重要成员, HCC中肿瘤源性外泌体被证实通过激活肿瘤微环境中的周围细胞参与促进癌细胞的转移[42]. 外泌体SNHG16可通过海绵吸附miR-4500, 通过PI3K/AKT/mTOR途径靶向GALNT1, 转运至血管内皮细胞发挥促血管生成作用[43], 导致HCC快速生长、早期转移等; 还可通过竞争性结合miR-942-3p促进间质细胞分泌MMP9, 同样发挥促进HCC转移的作用[44]. 以上多项研究提示: SNHG16在HCC发挥促癌作用. 然而, Xu等[45]的一项研究发现SNHG16在HCC组织和HCC细胞系中均较正常对照组表达显著下调. 过表达SNHG16抑制HCC细胞增殖、5-FU化疗耐药和肿瘤生长, 提示SNHG16可能作为肿瘤抑制因子抑制HCC的发展. 目前关于SNHG16在HCC中的作用(图1D), 研究者意见不一, 这可能与肿瘤特定微环境有关, 仍需后续进一步临床和实验研究.
综上所述, SNHG16在几乎所有消化道肿瘤中都被认为是一种致癌的lncRNA. SNHG16通过影响下游靶基因或经典信号通路的表达, 参与肿瘤细胞的增殖、迁移、凋亡, 通过调控肿瘤微环境介导转移, 通过"海绵化"miRNA介导肿瘤细胞耐药反应等, 目前已发现超过20种miRNA与SNHG16相互作用, 我们总结了消化道肿瘤中SNHG16与miRNA的海绵作用及功能研究(表1), 以及SNHG16的表达与肿瘤临床特征之间的相关性(表2). 尽管目前在消化道肿瘤的研究中提示, SNHG16主要作为ceRNA发挥作用, 但lncRNA机制复杂多样, 整体且详尽的调控机制有待进一步挖掘. 另外, 虽然研究证实在不同肿瘤类型的组织样本中SNHG16表达升高, 但尚未评估该lncRNA是否可以作为早期检测癌症的循环标志物. 因此, 探索SNHG16作为恶性肿瘤早期诊断的价值和意义以及开发新型靶向药物仍是我们未来努力的方向.
| 肿瘤类型 | 表达水平 | 致癌/抑癌基因 | 功能作用 | miRNAs | 目标基因 | 参考文献 |
| GC | Up-regulated | Oncogene | Proliferation, migration, invasion, apoptosis | / | / | [7] |
| GC | Up-regulated | Oncogene | Proliferation, invasionability, apoptosis | miR-135a-5p | JAK2/STAT3 | [8] |
| GC | Up-regulated | Oncogene | Mediating 5-fu resistance | miR-506-3p | PTBP1 | [9] |
| GC | Up-regulated | Oncogene | EMT, invasion | / | DKK3 | [10] |
| GC | Up-regulated | Oncogene | Proliferation, apoptosis, migration, invasion | / | P53, c-Myc | [11] |
| CRC | Up-regulated | Oncogene | Migration, apoptosis | / | ASCL2, ETS2, c-Myc | [13] |
| CRC | Up-regulated | Oncogene | Proliferation, migration | miR-200a-3p | / | [14] |
| Colon cancer | Up-regulated | Oncogene | Proliferation | miR-302a-3p | AKT | [15] |
| CRC | Up-regulated | Oncogene | Proliferation, migration, invasion, apoptosis | miR-132-3p | USP22 | [16] |
| CRC | Up-regulated | Oncogene | Proliferation, apoptosis | miR-214-3p | ABCB1 | [17] |
| CRC | Up-regulated | Oncogene | Proliferation, migration, invasion, EMT | miR-124-3p | MCP-1 | [18] |
| CRC | Up-regulated | Oncogene | Proliferation, migration, invasion, EMT | miR-195-5p | YAP1/TEAD1 | [19] |
| ESCC | Up-regulated | Oncogene | Proliferation, invasion, apoptosis | / | c-myc, cyclin | [21] |
| ESCC | Up-regulated | Oncogene | Proliferation, migration, apoptosis, EMT | miR-140-5p | ZEB1 | [22] |
| EC | Up-regulated | Oncogene | Proliferation | miR-802 | PTCH1 | [23] |
| ESCC | Up-regulated | Oncogene | Proliferation, migration | / | EIF4A3/RhoU | [24] |
| PC | Up-regulated | Oncogene | Proliferation, migration, invasion, apoptosis | miR-302b-3p | SLC2A4 | [26] |
| PC | Up-regulated | Oncogene | Proliferation, migration, invasion, lipogenesis | miR-195 | SREBP2 | [27] |
| CCA | Up-regulated | Oncogene | Proliferation, apoptosis | miR-146a-5p | GATA6 | [29] |
| HCC | Up-regulated | Oncogene | Proliferation, migration, invasion | / | / | [31] |
| HCC | Up-regulated | Oncogene | Proliferation, migration, invasion, apoptosis | miR-17-5p | P62 | [32] |
| HCC | Up-regulated | Oncogene | Invasiveness, proliferation | / | / | [33] |
| HCC | Up-regulated | Oncogene | Proliferation, migration, invasion | miR-186 | / | [34] |
| HCC | Up-regulated | Oncogene | Proliferation, invasion tumorigenesis | miR-195 | / | [35] |
| HCC | Up-regulated | Oncogene | Proliferation, migration, invasion, apoptosis and EMT | miR-4500 | STAT3 | [36] |
| HCC | Up-regulated | Oncogene | Proliferation | miR-302a-3p | FGF19 | [37] |
| HCC | Up-regulated | Oncogene | Metastasis, EMT | miR-605-3p | TRAF6、NF-κB | [38] |
| HCC | Up-regulated | Oncogene | Viability, apoptosis | miR-23b-3p | EGR1 | [39] |
| HCC | Up-regulated | Oncogene | Proliferation, migration, invasion, EMT, apoptosis | let-7b-5p | CDC25B、HMGA2 | [41] |
| HCC | Up-regulated | Oncogene | Angiogenesis | miR-4500 | GALNT1、PI3K/Akt/mTOR | [43] |
| HCC | Up-regulated | Oncogene | Metastasis | miR 942 3p | MMP9 | [44] |
| HCC | Down-regulated | Tumor suppressor | Proliferation, chemoresistance, tumorigenicity | hsa-miR-93 | / | [45] |
| 肿瘤类型 | 临床特征 | 参考文献 |
| GC | Invasion depth, lymph node metastasis, TNM stage, histological differentiation | [7] |
| GC | Tumor size, TNM stage, poor survival | [8] |
| GC | Invasion depth | [11] |
| CRC | TNM stage | [13] |
| CRC | Metastasis, lymph node | [14] |
| CRC | LV1, PNI, TNM stage, lymph node, metastasis, distant metastasis, poor prognosis | [19] |
| ESCC | Tumor stage, lymph nodes metastasis, clinical stage, poor survival, poor prognosis | [21] |
| ESCC | Tumour differentiation, T stage | [24] |
| PC | Poor prognosis | [26] |
| PC | Gemcitabine resistance | [28] |
| HCC | Tumor size, AFP level, PVTT, metastasis, poor survival, sorafenib resistance | [31] |
| HCC | Multiple tumors, macrovascular invasion, poor prognosis | [32] |
| HCC | Serum AFP level, poor prognosis | [33] |
| HCC | Tumor size, TNM stage, ALT level, HBV DNA level | [34] |
| HCC | Tumor size, lymph node status, TNM stage and poor prognosis | [36] |
| HCC | Poor prognosis | [37] |
| HCC | Tumour diameter, tumour thrombus, tumour satellites, poor prognosis | [38] |
| HCC | Sorafenib resistance | [39] |
| HCC | Tumor size, TNM stage, vascular invasion, sorafenib resistance | [40] |
| HCC | Poor prognosis | [44] |
| 1. | Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12-49. [PubMed] [DOI] |
| 2. | Feinberg AP, Levchenko A. Epigenetics as a mediator of plasticity in cancer. Science. 2023;379:eaaw3835. [PubMed] [DOI] |
| 3. | Wang N, Yu Y, Xu B, Zhang M, Li Q, Miao L. Pivotal prognostic and diagnostic role of the long noncoding RNA colon cancerassociated transcript 1 expression in human cancer (Review). Mol Med Rep. 2019;19:771-782. [PubMed] [DOI] |
| 4. | Yu M, Ohira M, Li Y, Niizuma H, Oo ML, Zhu Y, Ozaki T, Isogai E, Nakamura Y, Koda T, Oba S, Yu B, Nakagawara A. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol. 2009;34:931-938. [PubMed] [DOI] |
| 5. | Xiao Y, Xiao T, Ou W, Wu Z, Wu J, Tang J, Tian B, Zhou Y, Su M, Wang W. LncRNA SNHG16 as a potential biomarker and therapeutic target in human cancers. Biomark Res. 2020;8:41. [PubMed] [DOI] |
| 6. | Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci. 2020;21. [PubMed] [DOI] |
| 7. | Lian D, Amin B, Du D, Yan W. Enhanced expression of the long non-coding RNA SNHG16 contributes to gastric cancer progression and metastasis. Cancer Biomark. 2017;21:151-160. [PubMed] [DOI] |
| 8. | Wang X, Kan J, Han J, Zhang W, Bai L, Wu H. LncRNA SNHG16 Functions as an Oncogene by Sponging MiR-135a and Promotes JAK2/STAT3 Signal Pathway in Gastric Cancer. J Cancer. 2019;10:1013-1022. [PubMed] [DOI] |
| 9. | Ding Y, Gao S, Zheng J, Chen X. Blocking lncRNA-SNHG16 sensitizes gastric cancer cells to 5-Fu through targeting the miR-506-3p-PTBP1-mediated glucose metabolism. Cancer Metab. 2022;10:20. [PubMed] [DOI] |
| 10. | Zhou C, Zhao J, Liu J, Wei S, Xia Y, Xia W, Bi Y, Yan Z, Huang H. LncRNA SNHG16 promotes epithelial- mesenchymal transition via down-regulation of DKK3 in gastric cancer. Cancer Biomark. 2019;26:393-401. [PubMed] [DOI] |
| 11. | Zhao JJ, Liu JJ, Zhang YY, Xia Y, Du H, Yan ZQ, Zhou CH, Xia WS, Zellmer L, Liao DJ, Wei SX, Huang H. SNHG16 lncRNAs are overexpressed and may be oncogenic in human gastric cancer by regulating cell cycle progression. Neoplasma. 2022;69:49-58. [PubMed] [DOI] |
| 12. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [PubMed] [DOI] |
| 13. | Christensen LL, True K, Hamilton MP, Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen JB, Pedersen JS, Lund AH, Vang S, Stribolt K, Madsen MR, Laurberg S, McGuire SE, Ørntoft TF, Andersen CL. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. 2016;10:1266-1282. [PubMed] [DOI] |
| 14. | Li Y, Lu Y, Chen Y. Long non-coding RNA SNHG16 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer via sponging miR-200a-3p. Biosci Rep. 2019;39. [PubMed] [DOI] |
| 15. | Ke D, Wang Q, Ke S, Zou L, Wang Q. Long-Non Coding RNA SNHG16 Supports Colon Cancer Cell Growth by Modulating miR-302a-3p/AKT Axis. Pathol Oncol Res. 2020;26:1605-1613. [PubMed] [DOI] |
| 16. | He X, Ma J, Zhang M, Cui J, Yang H. Long Non-Coding RNA SNHG16 Activates USP22 Expression to Promote Colorectal Cancer Progression by Sponging miR-132-3p. Onco Targets Ther. 2020;13:4283-4294. [PubMed] [DOI] |
| 17. | Tan P, Xu M, Nie J, Qin J, Liu X, Sun H, Wang S, Pan Y. LncRNA SNHG16 promotes colorectal cancer proliferation by regulating ABCB1 expression through sponging miR-214-3p. J Biomed Res. 2022;36:231-241. [PubMed] [DOI] |
| 18. | Chen ZY, Wang XY, Yang YM, Wu MH, Yang L, Jiang DT, Cai H, Peng Y. LncRNA SNHG16 promotes colorectal cancer cell proliferation, migration, and epithelial-mesenchymal transition through miR-124-3p/MCP-1. Gene Ther. 2022;29:193-205. [PubMed] [DOI] |
| 19. | Xiang Z, Huang G, Wu H, He Q, Yang C, Dou R, Liu Q, Song J, Fang Y, Wang S, Xiong B. SNHG16 upregulation-induced positive feedback loop with YAP1/TEAD1 complex in Colorectal Cancer cell lines facilitates liver metastasis of colorectal cancer by modulating CTCs epithelial-mesenchymal transition. Int J Biol Sci. 2022;18:5291-5308. [PubMed] [DOI] |
| 20. | Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020;13:1010-1021. [PubMed] [DOI] |
| 21. | Han GH, Lu KJ, Wang P, Ye J, Ye YY, Huang JX. LncRNA SNHG16 predicts poor prognosis in ESCC and promotes cell proliferation and invasion by regulating Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:3795-3803. [PubMed] [DOI] |
| 22. | Zhang K, Chen J, Song H, Chen LB. SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget. 2018;9:1028-1040. [PubMed] [DOI] |
| 23. | Zhang L, Liang H, Zhang J, Yang Y, Ling X, Jiang H. Long Non-coding RNA SNHG16 Facilitates Esophageal Cancer Cell Proliferation and Self-renewal through the microRNA-802/PTCH1 Axis. Curr Med Chem. 2022;29:6084-6099. [PubMed] [DOI] |
| 24. | Ren L, Fang X, Shrestha SM, Ji Q, Ye H, Liang Y, Liu Y, Feng Y, Dong J, Shi R. LncRNA SNHG16 promotes development of oesophageal squamous cell carcinoma by interacting with EIF4A3 and modulating RhoU mRNA stability. Cell Mol Biol Lett. 2022;27:89. [PubMed] [DOI] |
| 25. | Zhao Z, Liu W. Pancreatic Cancer: A Review of Risk Factors, Diagnosis, and Treatment. Technol Cancer Res Treat. 2020;19:1533033820962117. [PubMed] [DOI] |
| 26. | Xu H, Miao X, Li X, Chen H, Zhang B, Zhou W. LncRNA SNHG16 contributes to tumor progression via the miR-302b-3p/SLC2A4 axis in pancreatic adenocarcinoma. Cancer Cell Int. 2021;21:51. [PubMed] [DOI] |
| 27. | Yu Y, Dong JT, He B, Zou YF, Li XS, Xi CH, Yu Y. LncRNA SNHG16 induces the SREBP2 to promote lipogenesis and enhance the progression of pancreatic cancer. Future Oncol. 2019;15:3831-3844. [PubMed] [DOI] |
| 28. | Yu Y, Zou YF, Hong RQ, Chen WJ, Chen L, Chen WQ, Wang HP, Yu Y. Long non-coding RNA SNHG16 decreased SMAD4 to induce gemcitabine resistance in pancreatic cancer via EZH2-mediated epigenetic modification. Kaohsiung J Med Sci. 2022;38:981-991. [PubMed] [DOI] |
| 29. | Wu T, Lei MS, Gao XZ, Xiong TG, Yang K, Gong Q, Tang R, Tian YP, Fu XH. lncRNA SNHG16 Mediates Cell Proliferation and Apoptosis in Cholangiocarcinoma by Directly Targeting miR-146a-5p/GATA6 Axis. Biochem Genet. 2021;59:1311-1325. [PubMed] [DOI] |
| 30. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [PubMed] [DOI] |
| 31. | Guo Z, Zhang J, Fan L, Liu J, Yu H, Li X, Sun G. Long Noncoding RNA (lncRNA) Small Nucleolar RNA Host Gene 16 (SNHG16) Predicts Poor Prognosis and Sorafenib Resistance in Hepatocellular Carcinoma. Med Sci Monit. 2019;25:2079-2086. [PubMed] [DOI] |
| 32. | Zhong JH, Xiang X, Wang YY, Liu X, Qi LN, Luo CP, Wei WE, You XM, Ma L, Xiang BD, Li LQ. The lncRNA SNHG16 affects prognosis in hepatocellular carcinoma by regulating p62 expression. J Cell Physiol. 2020;235:1090-1102. [PubMed] [DOI] |
| 33. | Zhang QJ, Li DZ, Lin BY, Geng L, Yang Z, Zheng SS. SNHG16 promotes hepatocellular carcinoma development via activating ECM receptor interaction pathway. Hepatobiliary Pancreat Dis Int. 2022;21:41-49. [PubMed] [DOI] |
| 34. | Chen H, Li M, Huang P. LncRNA SNHG16 Promotes Hepato-cellular Carcinoma Proliferation, Migration and Invasion by Regulating miR-186 Expression. J Cancer. 2019;10:3571-3581. [PubMed] [DOI] |
| 35. | Xie X, Xu X, Sun C, Yu Z. Long intergenic noncoding RNA SNHG16 interacts with miR-195 to promote proliferation, invasion and tumorigenesis in hepatocellular carcinoma. Exp Cell Res. 2019;383:111501. [PubMed] [DOI] |
| 36. | Lin Q, Zheng H, Xu J, Zhang F, Pan H. LncRNA SNHG16 aggravates tumorigenesis and development of hepatocellular carcinoma by sponging miR-4500 and targeting STAT3. J Cell Biochem. 2019;120:11604-11615. [PubMed] [DOI] |
| 37. | Li W, Xu W, Song JS, Wu T, Wang WX. LncRNA SNHG16 promotes cell proliferation through miR-302a-3p/FGF19 axis in hepatocellular carcinoma. Neoplasma. 2019;66:397-404. [PubMed] [DOI] |
| 38. | Hu YL, Feng Y, Chen YY, Liu JZ, Su Y, Li P, Huang H, Mao QS, Xue WJ. SNHG16/miR-605-3p/TRAF6/NF-κB feedback loop regulates hepatocellular carcinoma metastasis. J Cell Mol Med. 2020;24:7637-7651. [PubMed] [DOI] |
| 39. | Jing Z, Ye X, Ma X, Hu X, Yang W, Shi J, Chen G, Gong L. SNGH16 regulates cell autophagy to promote Sorafenib Resistance through suppressing miR-23b-3p via sponging EGR1 in hepatocellular carcinoma. Cancer Med. 2020;9:4324-4338. [PubMed] [DOI] |
| 40. | Ye J, Zhang R, Du X, Chai W, Zhou Q. Long noncoding RNA SNHG16 induces sorafenib resistance in hepatocellular carcinoma cells through sponging miR-140-5p. Onco Targets Ther. 2019;12:415-422. [PubMed] [DOI] |
| 41. | Li S, Peng F, Ning Y, Jiang P, Peng J, Ding X, Zhang J, Jiang T, Xiang S. SNHG16 as the miRNA let-7b-5p sponge facilitates the G2/M and epithelial-mesenchymal transition by regulating CDC25B and HMGA2 expression in hepatocellular carcinoma. J Cell Biochem. 2020;121:2543-2558. [PubMed] [DOI] |
| 42. | Li R, Wang Y, Zhang X, Feng M, Ma J, Li J, Yang X, Fang F, Xia Q, Zhang Z, Shang M, Jiang S. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol Cancer. 2019;18:18. [PubMed] [DOI] |
| 43. | Li S, Qi Y, Huang Y, Guo Y, Huang T, Jia L. Exosome-derived SNHG16 sponging miR-4500 activates HUVEC angiogenesis by targeting GALNT1 via PI3K/Akt/mTOR pathway in hepatocellular carcinoma. J Physiol Biochem. 2021;77:667-682. [PubMed] [DOI] |
| 44. | Xu Y, Luan G, Li Z, Liu Z, Qin G, Chu Y. Tumour-derived exosomal lncRNA SNHG16 induces telocytes to promote metastasis of hepatocellular carcinoma via the miR-942-3p/MMP9 axis. Cell Oncol (Dordr). 2023;46:251-264. [PubMed] [DOI] |
| 45. | Xu F, Zha G, Wu Y, Cai W, Ao J. Overexpressing lncRNA SNHG16 inhibited HCC proliferation and chemoresistance by functionally sponging hsa-miR-93. Onco Targets Ther. 2018;11:8855-8863. [PubMed] [DOI] |
学科分类: 胃肠病学和肝病学
手稿来源地: 江苏省
同行评议报告学术质量分类
A级 (优秀): 0
B级 (非常好): B
C级 (良好): C, C, C, C
D级 (一般): 0
E级 (差): 0
科学编辑: 张砚梁 制作编辑:张砚梁